Borrelia codon usage
Data
Report for BBA15
Codon Adaptation Index (CAI) for sequence : 0.47
GC percentage for sequence : 33.58%
GENETIC CODE USED : 1 <a href="http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi">more about genetic code</a>
CODON USAGE
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| COUNT | 1 | 5 | O | 7 | O | O | O | O | 2 | O | 6 | 7 | 11 | 7 | O | 1 | 4 | O | 4 | 19 | 10 | 4 | 2 | 6 | O | O | 8 | O | 5 | 11 | O | 9 | 5 | O | 3 | 4 | 39 | 2 | 1 | 2 | O | O | O | 1 | 4 | 10 | 6 | O | 1 | 6 | O | O | O | 13 | 14 | 3 | O | 1 | 2 | 3 | O | 2 | 11 | 11 |
Relative Synonimous Codon Usage (RSCU)
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| RSCU | 0.31 | 1.54 | 0.00 | 2.15 | 0.00 | 0.00 | 0.00 | 0.00 | 6.00 | 0.00 | 0.92 | 1.08 | 1.22 | 0.78 | 0.00 | 2.00 | 2.00 | 0.00 | 0.35 | 1.65 | 1.82 | 0.73 | 0.36 | 1.09 | 0.00 | 0.00 | 1.85 | 0.00 | 1.15 | 2.36 | 0.00 | 1.93 | 1.07 | 0.00 | 0.64 | 0.19 | 1.81 | 1.00 | 0.67 | 1.33 | 0.00 | 0.00 | 0.00 | 4.00 | 0.89 | 2.22 | 1.33 | 0.00 | 0.22 | 1.33 | 0.00 | 0.00 | 0.00 | 1.73 | 1.87 | 0.40 | 0.00 | 1.00 | 0.80 | 1.20 | 0.00 | 0.33 | 1.83 | 1.83 |
Relative Adaptiveness of Codon
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| RAC | 0.14 | 0.71 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.86 | 1.00 | 1.00 | 0.64 | 0.00 | 1.00 | 1.00 | 0.00 | 0.21 | 1.00 | 1.00 | 0.40 | 0.20 | 0.60 | 0.00 | 0.00 | 1.00 | 0.00 | 0.62 | 1.00 | 0.00 | 0.82 | 0.45 | 0.00 | 0.27 | 0.10 | 1.00 | 1.00 | 0.50 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.40 | 1.00 | 0.60 | 0.00 | 0.10 | 0.60 | 0.00 | 0.00 | 0.00 | 0.93 | 1.00 | 0.21 | 0.00 | 1.00 | 0.67 | 1.00 | 0.00 | 0.18 | 1.00 | 1.00 |
Monomers
| A | 352 |
| T | 192 |
| G | 154 |
| C | 121 |
<a href="http://search.cpan.org/~shardiwal/Bio-Tools-CodonOptTable-1.05/lib/Bio/Tools/CodonOptTable.pm">Source code</a> is available.
Report for BBA24
Codon Adaptation Index (CAI) for sequence : 0.50
GC percentage for sequence : 32.46%
GENETIC CODE USED : 1 <a href="http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi">more about genetic code</a>
CODON USAGE
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| COUNT | 1 | 7 | 1 | 7 | 2 | 1 | O | O | 2 | O | 4 | 8 | 2 | 7 | 2 | 2 | 4 | O | 3 | 15 | 5 | 2 | 1 | 1 | O | 1 | 5 | O | 7 | 7 | O | 5 | 3 | 1 | 1 | 3 | 24 | 5 | 6 | 2 | O | O | 3 | 1 | 3 | 5 | 1 | O | O | 1 | O | O | O | 8 | 8 | 1 | 1 | O | 1 | 1 | 2 | 2 | 2 | 4 |
Relative Synonimous Codon Usage (RSCU)
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| RSCU | 0.25 | 1.75 | 0.25 | 1.75 | 2.40 | 1.20 | 0.00 | 0.00 | 2.40 | 0.00 | 0.67 | 1.33 | 0.44 | 1.56 | 1.00 | 1.00 | 2.00 | 0.00 | 0.33 | 1.67 | 2.22 | 0.89 | 0.44 | 0.44 | 0.00 | 2.00 | 1.25 | 0.00 | 1.75 | 2.47 | 0.00 | 1.76 | 1.06 | 0.35 | 0.35 | 0.22 | 1.78 | 1.00 | 1.50 | 0.50 | 0.00 | 0.00 | 3.00 | 1.00 | 1.80 | 3.00 | 0.60 | 0.00 | 0.00 | 0.60 | 0.00 | 0.00 | 0.00 | 1.78 | 1.78 | 0.22 | 0.22 | 0.00 | 1.00 | 1.00 | 0.80 | 0.80 | 0.80 | 1.60 |
Relative Adaptiveness of Codon
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| RAC | 0.14 | 1.00 | 0.14 | 1.00 | 1.00 | 0.50 | 0.00 | 0.00 | 1.00 | 0.00 | 0.50 | 1.00 | 0.29 | 1.00 | 1.00 | 1.00 | 1.00 | 0.00 | 0.20 | 1.00 | 1.00 | 0.40 | 0.20 | 0.20 | 0.00 | 1.00 | 0.71 | 0.00 | 1.00 | 1.00 | 0.00 | 0.71 | 0.43 | 0.14 | 0.14 | 0.13 | 1.00 | 1.00 | 1.00 | 0.33 | 0.00 | 0.00 | 1.00 | 0.33 | 0.60 | 1.00 | 0.20 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 | 0.00 | 1.00 | 1.00 | 0.13 | 0.13 | 0.00 | 1.00 | 1.00 | 0.50 | 0.50 | 0.50 | 1.00 |
Monomers
| A | 248 |
| T | 139 |
| G | 102 |
| C | 84 |
<a href="http://search.cpan.org/~shardiwal/Bio-Tools-CodonOptTable-1.05/lib/Bio/Tools/CodonOptTable.pm">Source code</a> is available.
Report for BBA68
Codon Adaptation Index (CAI) for sequence : 0.48
GC percentage for sequence : 27.62%
GENETIC CODE USED : 1 <a href="http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi">more about genetic code</a>
CODON USAGE
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| COUNT | 2 | 6 | O | 3 | O | O | O | O | 3 | O | 4 | 16 | 1 | 12 | 2 | O | 13 | O | 4 | 21 | 3 | 2 | 1 | 1 | 2 | 1 | 12 | 5 | 14 | 7 | 2 | 9 | 5 | O | 3 | 4 | 33 | 4 | 7 | 2 | 1 | O | 1 | 4 | 1 | 5 | 3 | O | 2 | 4 | O | O | O | 8 | O | 4 | 1 | 1 | 3 | 6 | O | O | 2 | 1 |
Relative Synonimous Codon Usage (RSCU)
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| RSCU | 0.73 | 2.18 | 0.00 | 1.09 | 0.00 | 0.00 | 0.00 | 0.00 | 6.00 | 0.00 | 0.40 | 1.60 | 0.15 | 1.85 | 2.00 | 0.00 | 2.00 | 0.00 | 0.32 | 1.68 | 1.71 | 1.14 | 0.57 | 0.57 | 1.33 | 0.67 | 1.16 | 0.48 | 1.35 | 1.62 | 0.46 | 2.08 | 1.15 | 0.00 | 0.69 | 0.22 | 1.78 | 1.00 | 1.56 | 0.44 | 0.67 | 0.00 | 0.67 | 2.67 | 0.40 | 2.00 | 1.20 | 0.00 | 0.80 | 1.60 | 0.00 | 0.00 | 0.00 | 2.46 | 0.00 | 1.23 | 0.31 | 1.00 | 0.67 | 1.33 | 0.00 | 0.00 | 2.67 | 1.33 |
Relative Adaptiveness of Codon
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| RAC | 0.33 | 1.00 | 0.00 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.25 | 1.00 | 0.08 | 1.00 | 1.00 | 0.00 | 1.00 | 0.00 | 0.19 | 1.00 | 1.00 | 0.67 | 0.33 | 0.33 | 1.00 | 0.50 | 0.86 | 0.36 | 1.00 | 0.78 | 0.22 | 1.00 | 0.56 | 0.00 | 0.33 | 0.12 | 1.00 | 1.00 | 1.00 | 0.29 | 0.25 | 0.00 | 0.25 | 1.00 | 0.20 | 1.00 | 0.60 | 0.00 | 0.40 | 0.80 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.50 | 0.13 | 1.00 | 0.50 | 1.00 | 0.00 | 0.00 | 1.00 | 0.50 |
Monomers
| A | 346 |
| T | 199 |
| G | 97 |
| C | 111 |
<a href="http://search.cpan.org/~shardiwal/Bio-Tools-CodonOptTable-1.05/lib/Bio/Tools/CodonOptTable.pm">Source code</a> is available.
Report for BBB18
Codon Adaptation Index (CAI) for sequence : 0.50
GC percentage for sequence : 29.17%
GENETIC CODE USED : 1 <a href="http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi">more about genetic code</a>
CODON USAGE
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| COUNT | 1 | 6 | O | 9 | O | 2 | 1 | 1 | 11 | 1 | 5 | 24 | 7 | 15 | 3 | 4 | 21 | 1 | 3 | 36 | 15 | 5 | 3 | 7 | 1 | 6 | 29 | 7 | 30 | 10 | O | 20 | 11 | 2 | 10 | 10 | 42 | 9 | 19 | 4 | O | 1 | 6 | 11 | 7 | 9 | 4 | O | 1 | 20 | O | O | O | 10 | 9 | 4 | O | 3 | 17 | 5 | 1 | 2 | 13 | 14 |
Relative Synonimous Codon Usage (RSCU)
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| RSCU | 0.25 | 1.50 | 0.00 | 2.25 | 0.00 | 0.75 | 0.38 | 0.38 | 4.13 | 0.38 | 0.34 | 1.66 | 0.64 | 1.36 | 0.86 | 1.14 | 1.91 | 0.09 | 0.15 | 1.85 | 2.00 | 0.67 | 0.40 | 0.93 | 0.29 | 1.71 | 1.32 | 0.32 | 1.36 | 1.13 | 0.00 | 2.26 | 1.25 | 0.23 | 1.13 | 0.38 | 1.62 | 1.00 | 1.65 | 0.35 | 0.00 | 0.22 | 1.33 | 2.44 | 1.02 | 1.32 | 0.59 | 0.00 | 0.15 | 2.93 | 0.00 | 0.00 | 0.00 | 1.74 | 1.57 | 0.70 | 0.00 | 1.00 | 1.55 | 0.45 | 0.13 | 0.27 | 1.73 | 1.87 |
Relative Adaptiveness of Codon
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| RAC | 0.11 | 0.67 | 0.00 | 1.00 | 0.00 | 0.18 | 0.09 | 0.09 | 1.00 | 0.09 | 0.21 | 1.00 | 0.47 | 1.00 | 0.75 | 1.00 | 1.00 | 0.05 | 0.08 | 1.00 | 1.00 | 0.33 | 0.20 | 0.47 | 0.17 | 1.00 | 0.97 | 0.23 | 1.00 | 0.50 | 0.00 | 1.00 | 0.55 | 0.10 | 0.50 | 0.24 | 1.00 | 1.00 | 1.00 | 0.21 | 0.00 | 0.09 | 0.55 | 1.00 | 0.35 | 0.45 | 0.20 | 0.00 | 0.05 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.90 | 0.40 | 0.00 | 1.00 | 1.00 | 0.29 | 0.07 | 0.14 | 0.93 | 1.00 |
Monomers
| A | 626 |
| T | 496 |
| G | 248 |
| C | 214 |
<a href="http://search.cpan.org/~shardiwal/Bio-Tools-CodonOptTable-1.05/lib/Bio/Tools/CodonOptTable.pm">Source code</a> is available.
Report for Whole Genome
Codon Adaptation Index (CAI) for sequence : 0.57
GC percentage for sequence : 28.86%
GENETIC CODE USED : 1 <a href="http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi">more about genetic code</a>
CODON USAGE
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| COUNT | 2000 | 7080 | 1187 | 8110 | 1277 | 3427 | 913 | 512 | 11465 | 584 | 6857 | 25195 | 3762 | 15623 | 2397 | 3330 | 9874 | 2224 | 6834 | 19297 | 8360 | 5151 | 3452 | 3285 | 1808 | 4294 | 23760 | 4165 | 17140 | 11759 | 1426 | 17941 | 4742 | 1385 | 8043 | 11148 | 37595 | 7794 | 24077 | 3813 | 648 | 1607 | 3746 | 4146 | 6834 | 7372 | 5162 | 1072 | 1834 | 9876 | 2607 | 3902 | 6838 | 6995 | 8161 | 2444 | 1195 | 3298 | 15345 | 4760 | 1204 | 2398 | 11350 | 6016 |
Relative Synonimous Codon Usage (RSCU)
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| RSCU | 0.44 | 1.54 | 0.26 | 1.77 | 0.42 | 1.13 | 0.30 | 0.17 | 3.78 | 0.19 | 0.43 | 1.57 | 0.39 | 1.61 | 0.84 | 1.16 | 1.63 | 0.37 | 0.52 | 1.48 | 1.65 | 1.02 | 0.68 | 0.65 | 0.59 | 1.41 | 1.58 | 0.28 | 1.14 | 1.56 | 0.19 | 2.38 | 0.63 | 0.18 | 1.07 | 0.46 | 1.54 | 1.00 | 1.73 | 0.27 | 0.26 | 0.63 | 1.48 | 1.63 | 1.28 | 1.38 | 0.96 | 0.20 | 0.34 | 1.84 | 0.59 | 0.88 | 1.54 | 1.49 | 1.74 | 0.52 | 0.25 | 1.00 | 1.53 | 0.47 | 0.23 | 0.46 | 2.17 | 1.15 |
Relative Adaptiveness of Codon
| CODON | GCC | GCA | GCG | GCT | CGA | AGG | CGT | CGG | AGA | CGC | AAC | AAT | GAC | GAT | TGC | TGT | CAA | CAG | GAG | GAA | GGA | GGT | GGG | GGC | CAC | CAT | ATT | ATC | ATA | CTT | CTG | TTA | CTA | CTC | TTG | AAG | AAA | ATG | TTT | TTC | CCG | CCC | CCA | CCT | AGT | TCA | AGC | TCG | TCC | TCT | TAG | TGA | TAA | ACT | ACA | ACC | ACG | TGG | TAT | TAC | GTC | GTG | GTT | GTA |
| AMINOACID | Ala | Ala | Ala | Ala | Arg | Arg | Arg | Arg | Arg | Arg | Asn | Asn | Asp | Asp | Cys | Cys | Gln | Gln | Glu | Glu | Gly | Gly | Gly | Gly | His | His | Ile | Ile | Ile | Leu | Leu | Leu | Leu | Leu | Leu | Lys | Lys | Met | Phe | Phe | Pro | Pro | Pro | Pro | Ser | Ser | Ser | Ser | Ser | Ser | Stop | Stop | Stop | Thr | Thr | Thr | Thr | Trp | Tyr | Tyr | Val | Val | Val | Val |
| RAC | 0.25 | 0.87 | 0.15 | 1.00 | 0.11 | 0.30 | 0.08 | 0.04 | 1.00 | 0.05 | 0.27 | 1.00 | 0.24 | 1.00 | 0.72 | 1.00 | 1.00 | 0.23 | 0.35 | 1.00 | 1.00 | 0.62 | 0.41 | 0.39 | 0.42 | 1.00 | 1.00 | 0.18 | 0.72 | 0.66 | 0.08 | 1.00 | 0.26 | 0.08 | 0.45 | 0.30 | 1.00 | 1.00 | 1.00 | 0.16 | 0.16 | 0.39 | 0.90 | 1.00 | 0.69 | 0.75 | 0.52 | 0.11 | 0.19 | 1.00 | 0.38 | 0.57 | 1.00 | 0.86 | 1.00 | 0.30 | 0.15 | 1.00 | 1.00 | 0.31 | 0.11 | 0.21 | 1.00 | 0.53 |
Monomers
| A | 524241 |
| T | 440303 |
| G | 225741 |
| C | 165502 |
<a href="http://search.cpan.org/~shardiwal/Bio-Tools-CodonOptTable-1.05/lib/Bio/Tools/CodonOptTable.pm">Source code</a> is available.
Charts
| BBA15 | 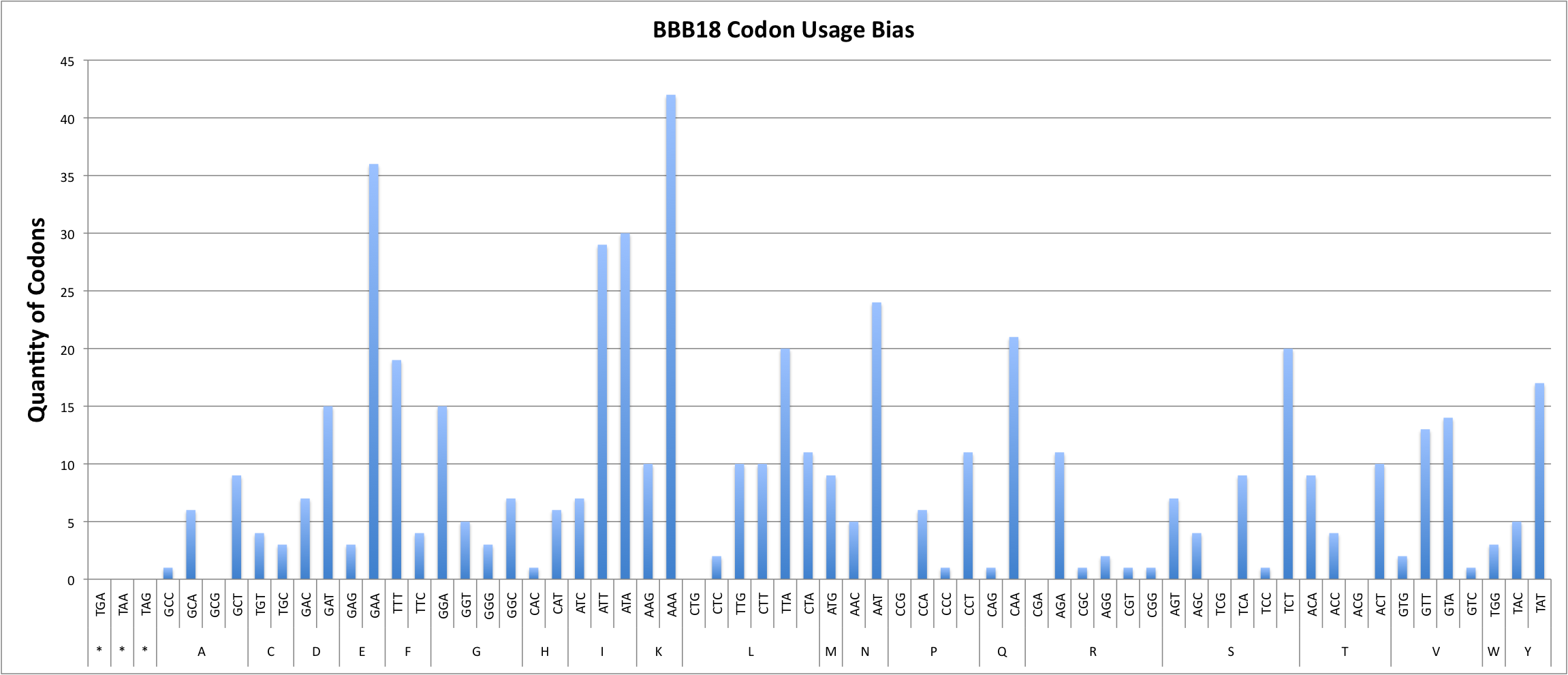
|
| BBA24 | 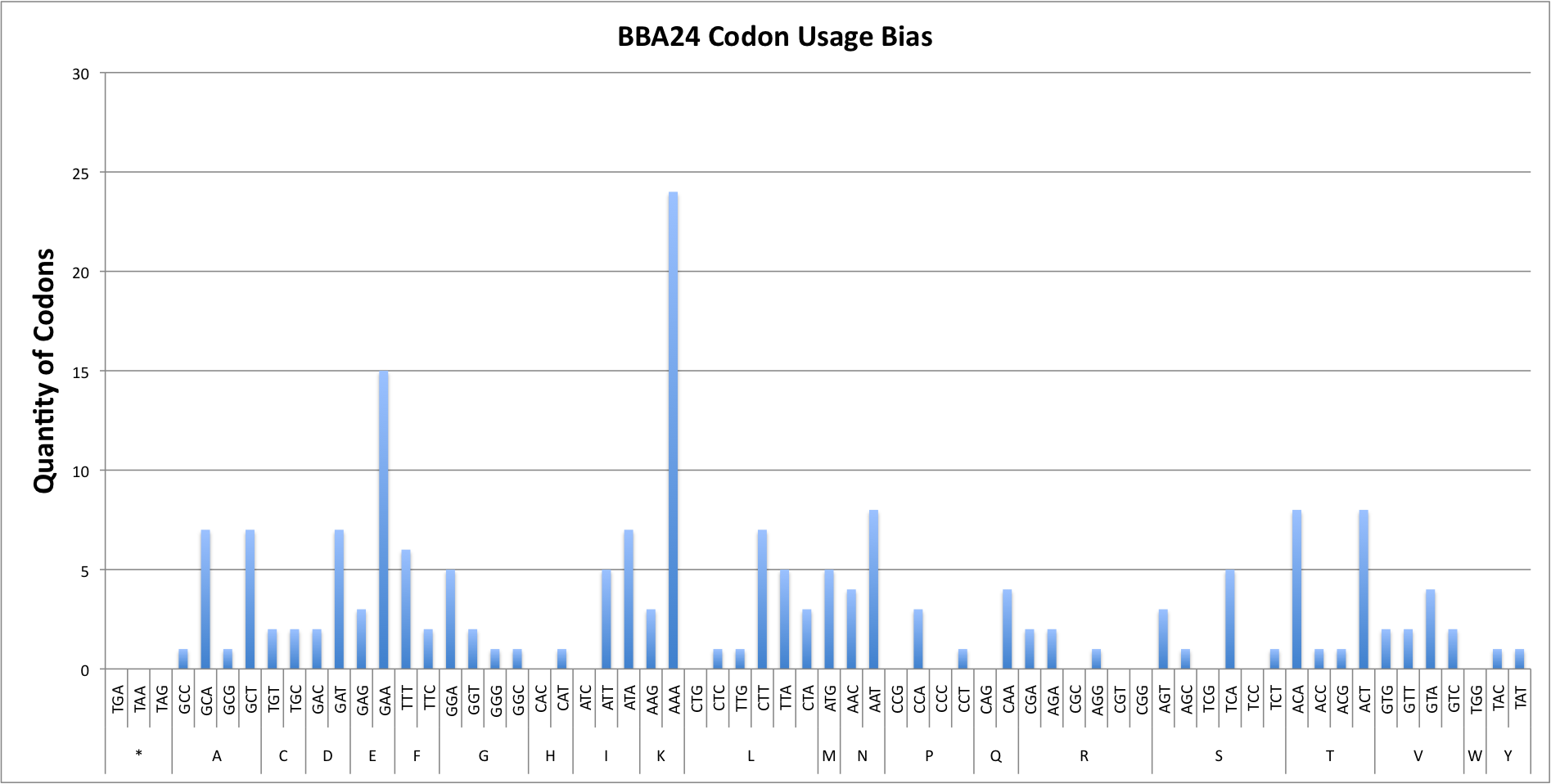
|
| BBA68 | 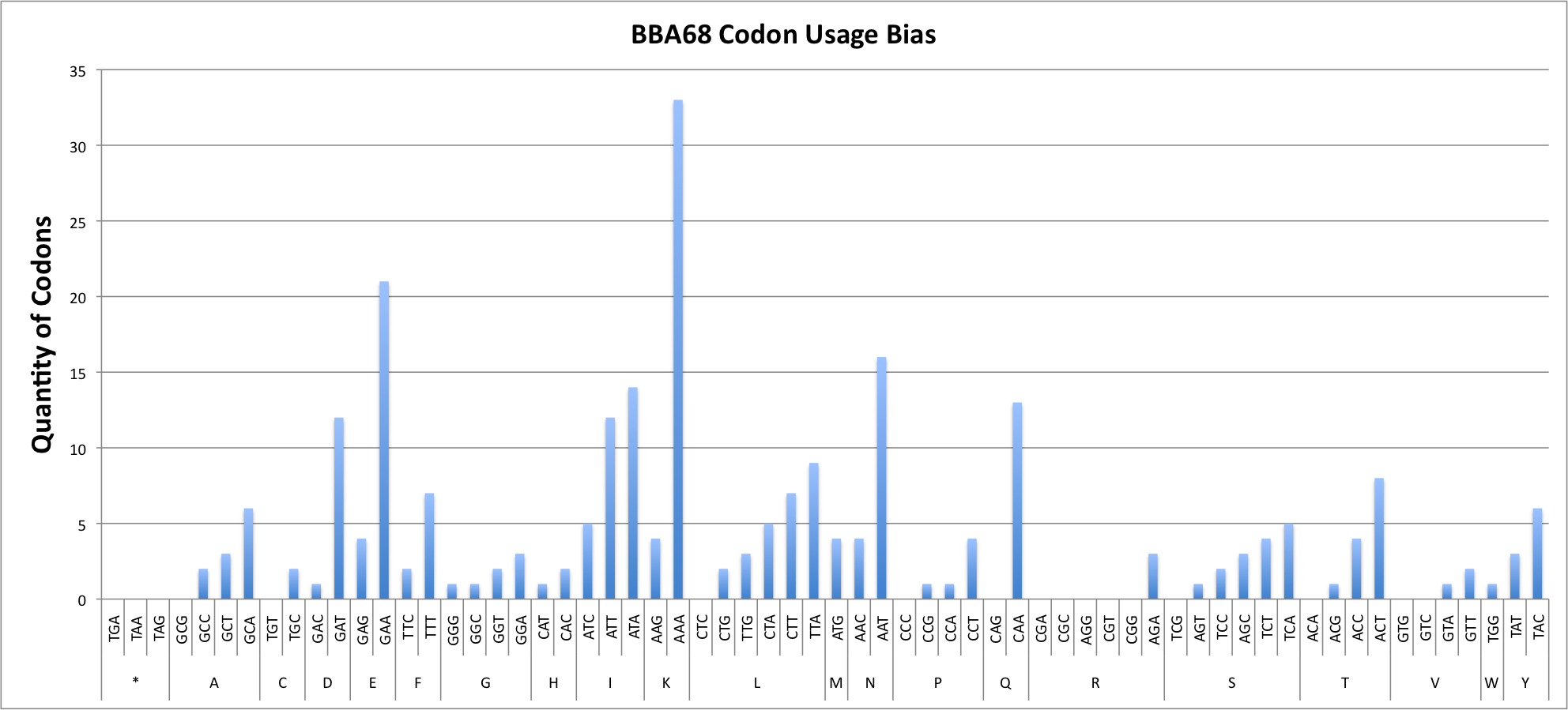
|
| BBB 18 | 
|
| Whole genome | 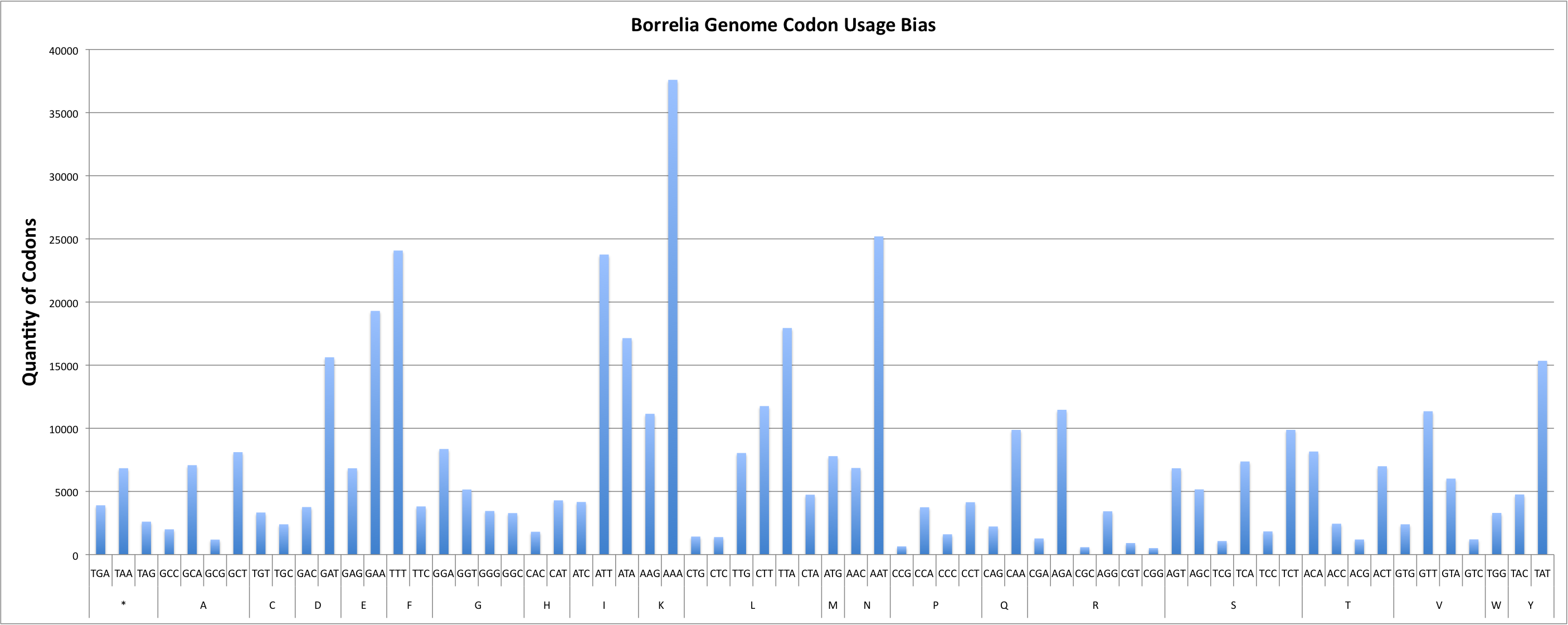
|
Hypothesis & Background
- Central Hypothesis: Borrelia host-interacting genes show optimal codon usage
- Background/Rationale:
- Borrelia is an obligate, non-free-living parasite of vertebrates. A large number of genes are devoted to host invasion and to surviving the host immune defense.
- Evolutionary theory predicts that highly expressed genes use the most abundant tRNA in the cell, and, as a result, they tend to show strong codon usage biases
- We expect Borrelia host-interacting, virulence-conferring genes use more optimal codons than housekeeping genes
- Importance: If the hypothesis is supported, it would establish a new computational method to identify host-interacting virulence genes based on genome analysis.
Data & Overview
- Data Set: The Borrelia burgdorferi B31 genome sequences, N=1500 genes
- Identify host-interacting genes and a set of housekeeping genes
- Calculate the Codon Usage Adaptation Index for each gene
- Test if the biases are significantly different between the virulence genes vs the house keeping genes
- Presentation, Report, & Future directions
Essential Computing skills
- Operating System: Linux/Ubuntu
- Programming Languages: BASH, Perl/BioPerl
- Database Language: SQL
- Statistical Language: R
Essential Readings
- Note: I will post some of the PDFs later
- Borrelia burgdorferi & Lyme disease: A Review
- Codon Usage Biases: Wikipedia
- Borrelia gene regulation: A Review
- Identification of Borrelia virulence genes: A genome-wide expression study
Weigang 11:38, 10 May 2013 (EDT)