Bioinformatics Workshop 2014
Course Description
Background
Biomedical research is becoming a high-throughput science. As a result, information technology plays an increasingly important role in biomedical discovery. Bioinformatics is a new interdisciplinary field formed by the merging of molecular biology and computer science techniques.Today’s biology students must therefore not only learn to perform in vivo and invitro, but also in silico research skills. Quantitative/computational biologists are expected to be in increasing demand in the 21st century.
However, the technical barrier to enter the field and perform basic research projects in a bioinformatics lab is daunting for most undergraduate students. This is mainly due to the multidisciplinary nature of quantitative biology, which requires understandings and skills in chemistry, biology, computer programming, and statistics. The Hunter Summer Bioinformatics Workshop aims to introduce bioinformatics to motivated undergraduate and high school students by lowering the barrier and dispensing the usual pre-requisites in advanced biology/chemistry courses as well as entry-level programming/statistics courses. The Workshop does not assume prior programming experience.
The workshop DOES NOT
- Replace existing advanced bioinformatics courses such as BIOL425 and STAT 319
- Teach advanced bioinformatics programming skills (e.g., advanced data structure, object-oriented Perl, BioPerl, or relational database with SQL), which are the contents of BIOL425
- Teach in-depth statistics or the popular R statistical package, although probabilistic thinking (e.g., distributions of a random variable, stochastic processes, likelihood, clustering analysis) is at the core of all bioinformatics analysis (STAT 319 teaches these topics)
To learn these advanced bioinformatics topics and skills, motivated students are encouraged to enroll in one of the Five Bioinformatics Concentrations of at Hunter. The QuBi program prepares the students for bioinformatics positions in a research lab or a biotechnology company.
Contents
This course will introduce both bioinformatics theories and practices. Topics include: database searching, sequence alignment, and basic molecular phylogenetics. The course is held in a UNIX-based instructional lab specifically configured for bioinformatics applications. Each session consists of a first-half instruction on bioinformatics theories and a second-half session of hands-on exercises.
Learning Goals
Students are expected to be able to:
- Retrieve and analyze DNA and protein sequences using online databases
- Write simple computer programs to manipulate DNA sequences
Textbook
No textbook required, handouts will be provided in the class.
Grading & Academic Honesty
Hunter College regards acts of academic dishonesty (e.g., plagiarism, cheating on examinations, obtaining unfair advantage, and falsification of records and official documents) as serious offenses against the values of intellectual honesty. The College is committed to enforcing the CUNY Policy on Academic Integrity and will pursue cases of academic dishonesty according to the Hunter College Academic Integrity Procedures.
Student performance will be evaluated by weekly assignments and projects. While these are take-home projects and students are allowed to work in groups, students are expected to compose the final short answers, computer commands, and code independently. There are virtually an unlimited number of ways to solve a computational problem, as are ways and personal styles to implement an algorithm. Writings and blocks of codes that are virtually exact copies between individual students will be investigated as possible cases of plagiarism (e.g., copies from the Internet, text book, or each other). In such a case, the instructor will hold closed-door exams for involved individuals. Zero credits will be given to ALL involved individuals if the instructor considers there is enough evidence for plagiarism. To avoid being investigated for plagiarism, Do Not Copy from Others & Do Not Let Others Copy Your Work.
The grading scheme for the course, is as follows (Subject to some change. You will be notified with sufficient time):
- In-Class Assignments: 8 exercises, 20 points each. [Attendance is mandatory)
- Weekly assignment: 4 exercises, 10 points each
- Mid-term: 50 points, on July 24
- Final exam: 50 points, on August 14
Programming Assignment Expectations
All code must begin with the lines in the Perl slides, without exception. For each assignment, unless otherwise stated, I would like the full text of the source code. Since you cannot print using the text editor in the lab (even if you are connected from home), you must copy and paste the code into a word processor or a local text editor. If you are using a word processor, change the font to a fixed-width/monospace font. On Windows, this is usually Courier.
Code indentation is your personal taste, so long as it is consistent and readable. Use comments whenever you think either the code is unclear, or simply as a guideline for yourself. Well-commented code improves readability, but be careful not overdo it.
Also, unless otherwise stated, both the input and the output of the program must be submitted as well. This should also be in fixed-width font, and you should label it in such a way so that I know it is the program's input/output. This is so that I know that you've run the program, what data you have used, and what the program produced.
If you are working from the lab, one option is to email the code to yourself, change the font, and then print it somewhere else as there is no printer in the lab.
Course Schedule (Tuesdays and Thursdays)
July 15. Course Overview & Lab Setup
- Course Overview
- LECTURE SLIDES (Bioinformatics)
- WORKSHOP SLIDES:(Workshop1)
- Workshop on Linux proficiency:
- Terminal & the bash shell
- Text editing
- First program
| Assignment #1 DUE: July 22 |
|---|
| Read (What is Bioinformatics, public via arXiv [[1]]) |
| Required reading (Expression of Genetic Information, public via NCBI bookshelf [[2]]) |
Bioinformatics questions
|
Linux proficiency tests
|
July 17. The Central Dogma of Molecular Biology
- LECTURE SLIDES (Genes, Proteins, Mutations)
- WORKSHOP SLIDES:(Workshop 2) with corrections July 22
- Workshop on Linux proficiency:
- Managing files with bash commands
- Editing with vi
- Writing programs in Perl
July 22. Sequence alignment & homology searching with BLAST
- LECTURE SLIDES:(Lecture 3: Gene Alignments and Homology)
- READING MATERIAL:(Reading material on sequence alignments and BLAST algorithm)
- WORKSHOP SLIDES:(Workshop 3)
- Workshop on Linux proficiency:
- Input/Output with bash
- Perl Data
- Perl Input/Output
| Assignment #2 DUE: July 29 |
|---|
| Bioinformatics questions |
| Linux proficiency tests Some diseases have animal models which are useful for studying the disease of interest. Use NCBI's BLAST tool to help choose between two common laboratory animals for PSEN1.
Answer the following:
Download a table of hit results by selecting the "Download" widget near the top of the browser window. Select "Hit Table (csv)" and save it. To make this file readable, change the commas into tabs, by using the tr command. Research how to use the tr command to reformat the file and save the output to a new file. Finally, use cat with the -n option on the new file in order to number the lines and name it hit_results.txt. Answer or show the following:
|
- July 23 ANNOUNCEMENT: Current participants are invited to join the Google Group for help, questions, and discussion. Send an email to biowork2014+subscribe@googlegroups.com or visit https://groups.google.com/d/forum/biowork2014. IT IS VERY IMPORTANT that you GET MEMBERSHIP in this group. We will use it for answering questions over the weekend for the midterm.
July 24. Alignment and Phylogenetics
- LECTURE SLIDES:(Lecture 4: Alignments, Homology, Molecular Phylogenetics)
- REQUIRED READING:((Not) for the faint of the heart: Molecular Phylogenentics)
- WORKSHOP SLIDES:(Workshop 4)
- Workshop on Linux proficiency:
- Remote access
- Accounts and passwords
- File transfers
July 29. Structure of human genome & genes
- Lecture & Workshop Slides: Gene Genome Structure Powerpoint or PDF PDF updated with animation slides
- Perl Workshop Slides: Workshop 5
- Tree-thinking Quizzes: Baum etal 2005 Quiz PDF
July 31. Macro-evolution: Cross-species comparisons
- Learning Goal: Cross-species comparisons
- Perl & bash Workshop Slides: Workshop 6
- Web Exercise 1. Cross-species comparisons with HomoloGene
- From the NCBI "TAS2R38" Gene page, click "HomoloGene" link under the "Related Information" (right-side navigation panel)
- You should see a page listing TAS2R38 orthologous (i.e., same gene in different species) genes from 7 mammalian species, including human (Homo sapiens), chimpanzee (Pan troglodytes), macaque (Macaca mulatta), dog (Canis lupus familiaris), cow (Bos taurus), rat (Rattus norvegicus), and mouse (Mus musculus).
- Write down your expectations for the following species relationships:
- Is chimpanzee more closely related to macaque or to human?
- Is dog more related to mouse or to cow?
- Is rat and mouse more closely related than human and chimpanzee?
- Click on the link "Show Pairwise Alignment Scores" under "Protein Alignments" and fill in the following table when the page loads. Do these sequence-comparison results change your expectations in the above? Explain.
| Species pair | % Protein Sequence Differences | % DNA Seq Differences |
|---|---|---|
| Chimp-Human | ? | ? |
| Chimp-Macaque | ? | ? |
| Dog-Cow | ? | ? |
| Dog-Mouse | ? | ? |
| Rat-Mouse | ? | ? |
You can find exact differences by clicking on "Blast" for each pairwise comparisons.
- Movie Break: Origin of Species: Lizards in an Evolutionary Tree
- What are the two hypotheses explaining the origin of different ecomorphs of lizards on Caribbean Islands?
- What is the expected phylogeny under each hypothesis?
- Which hypothesis is supported by the phylogeny of actual DNA sequences?
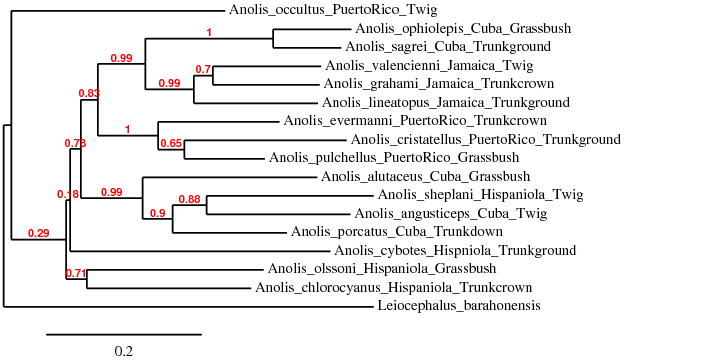
- Web Exercise 2. Derive your own tree of Anole lizards
- Copy and paste the lizard DNA sequences into a text editor. Using underscores, attach geographic location and ecomorph of each species according to the lizard card
- Go to the the phylogeny.fr web and select "Phylogenetic Analysis" and then "One Click" analysis
- Copy and paste your edited sequences into the text box and click on "Submit"
- When analysis is finished, you should see a phylogenetic tree. Answer the following two questions:
- Are species grouped by geography or by habitats?
- Which hypothesis is supported by your phylogenetic tree?
| Assignment #3 DUE: Aug 5 |
|---|
Bioinformatics questions
|
| Linux proficiency tests Scenario: Your research lab is interested in the 5' and 3' UTR regions of the human alpha-globin gene. You have been asked to take the same sequence file from above and prepare it for analysis. You have a Perl script that another lab member had used for data quality checks on another gene. Your script will need to transform the UTR bases from uppercase to lowercase. Another member prepared a control sequence in a FASTA file.
|
Aug 5. Micro-evolution: Human genetic variations
- Lecture Slides: Genetic Variation (Powerpoint) Slides updated
- Workshop Slides: Algorithm Thinking (Workshop 7)
- Short Film: Natural Selection in Humans
- Worksheets
| Assignment #4 DUE: Aug 12 |
|---|
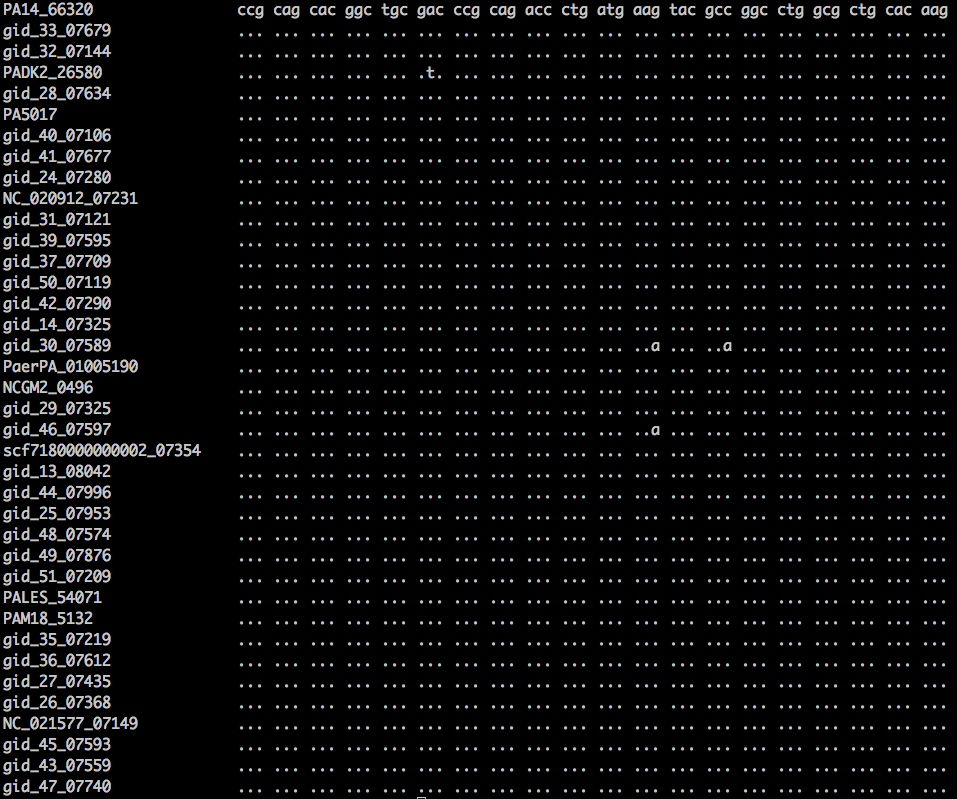
| Bioinformatics questions The image at left shows an alignment of 20 codons from 38 strains of a bacterial species. Answer the following questions:
|
Linux proficiency tests
|
Aug 7. Genome function: Transcriptome analysis
- Lecture Slides: Gene Expression (Powerpoint or PDF)
- In-Class Exercise
- Read this experimental report and extract the following information:
- Name of the two species used in experiments
- How many genes were measured for their expression (i.e., mRNA) levels?
- Describe a biological question that can be answered by this experiment (e.g., which genes are expressed at a particular developmental stage)
- Go to dictyExpress and explore the time course of a set of genes
- Choose the 2nd Box: "Run dictyExpress (RNA-seq)"
- In the "Gene Selection" Box, type the following gene names one at a time (DON'T copy and paste; when the gene is found, highlight it and press enter): acrA, catB, dcsA, acgA, abcG18
- Click "Update" and answer the question based on the plot in the "Expression Profile" panel: Are these genes up- or down-regulated during development?
- Do the same for the 2nd set of genes: mserS, rpl38, rpsA, rpl35a, gfm1
- Do the same for the 3rd set of genes: gefB, gefX, gxcB, mgp3, gefN
- Combine all 3 sets of genes and produce a heatmap
- In the "Hierarchical Clustering" Panel, choose the "Pearson Correlation" for "Distance Function"
- Choose "Average Linkage" for "Linkage" and your choice of color gradient
- What is represented by each row?
- What is represented by each column?
- Do these 3 sets of genes form clusters by themselves?
- HHMI slides: A technical description of how to group genes and samples by their overall similarity in gene expression levels
- Workshop Slides: Workshop 8 PDF
Aug 12. Review
- Submit teacher's evaluation:
- Computer: Submit using computer
- Smartphone: Submit using mobile
- Review Slides: Final Review (Powerpoint or PDF) and Informatics (PDF)