EEB BootCamp 2020: Difference between revisions
Jump to navigation
Jump to search
Bioinformatics Boot Camp for Ecology & Evolution: Pathogen Evolutionary Genomics
Thursday, Aug 6, 2020, 2 - 3:30pm
Instructors: Dr Weigang Qiu & Ms Saymon Akther
Email: weigang@genectr.hunter.cuny.edu
Lab Website: http://diverge.hunter.cuny.edu/labwiki/
imported>Weigang (Created page with "<center>Bioinformatics Boot Camp for Ecology & Evolution: '''Pathogen Evolutionary Genomics'''</center> <center>Thursday, Aug 6, 2020, 2 - 3:30pm</center> <center>'''Instructo...") |
imported>Weigang m (→Tutorial) |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 19: | Line 19: | ||
==Case studies from Qiu Lab== | ==Case studies from Qiu Lab== | ||
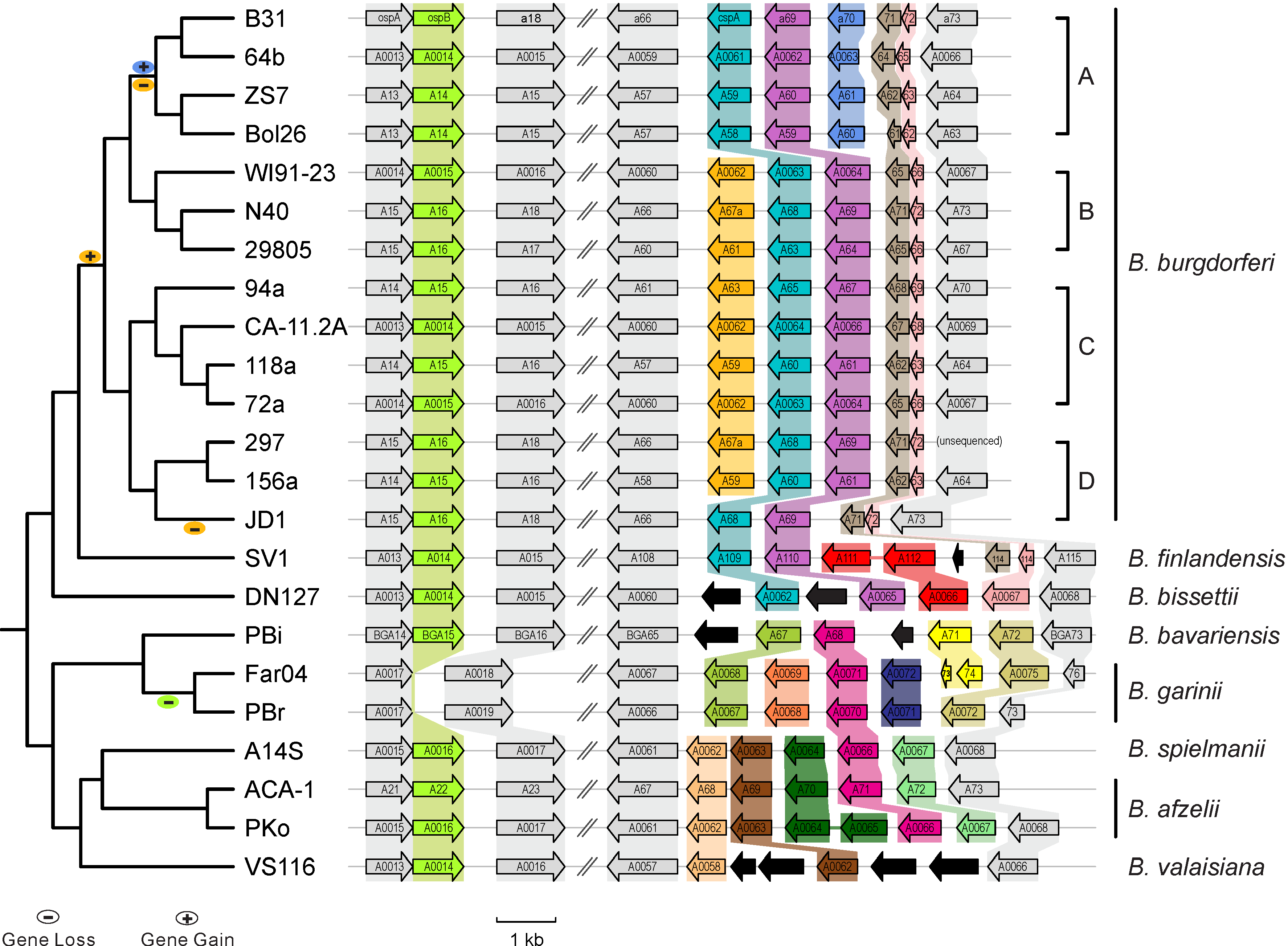
* [http://borreliabase.org Comparative genomics of worldwide Lyme disease pathogens] | * [http://borreliabase.org Comparative genomics of worldwide Lyme disease pathogens] | ||
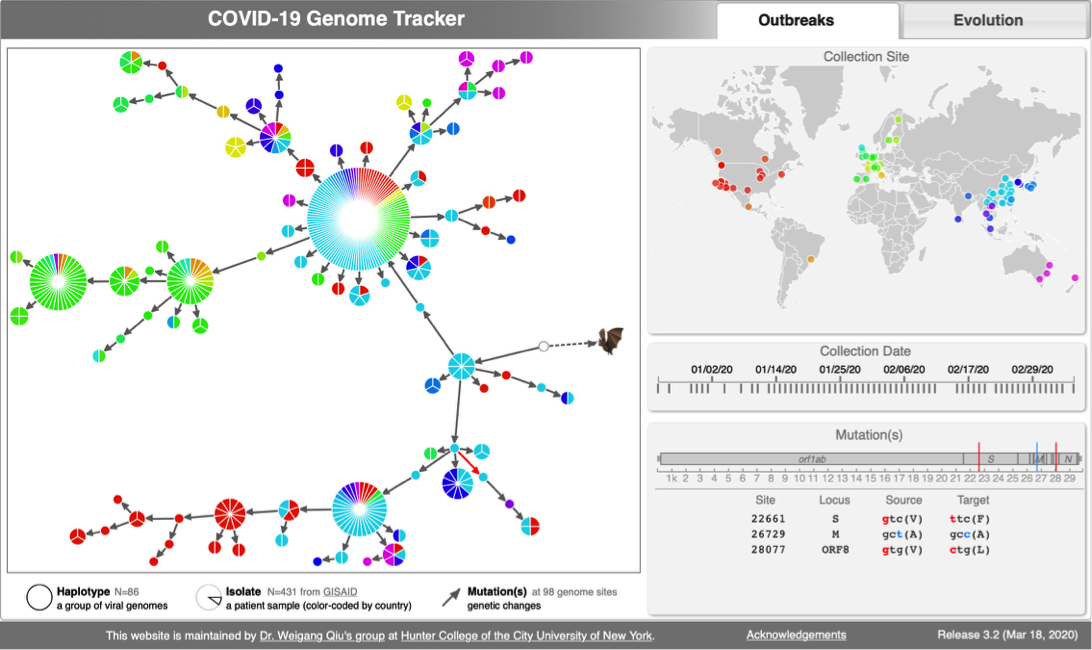
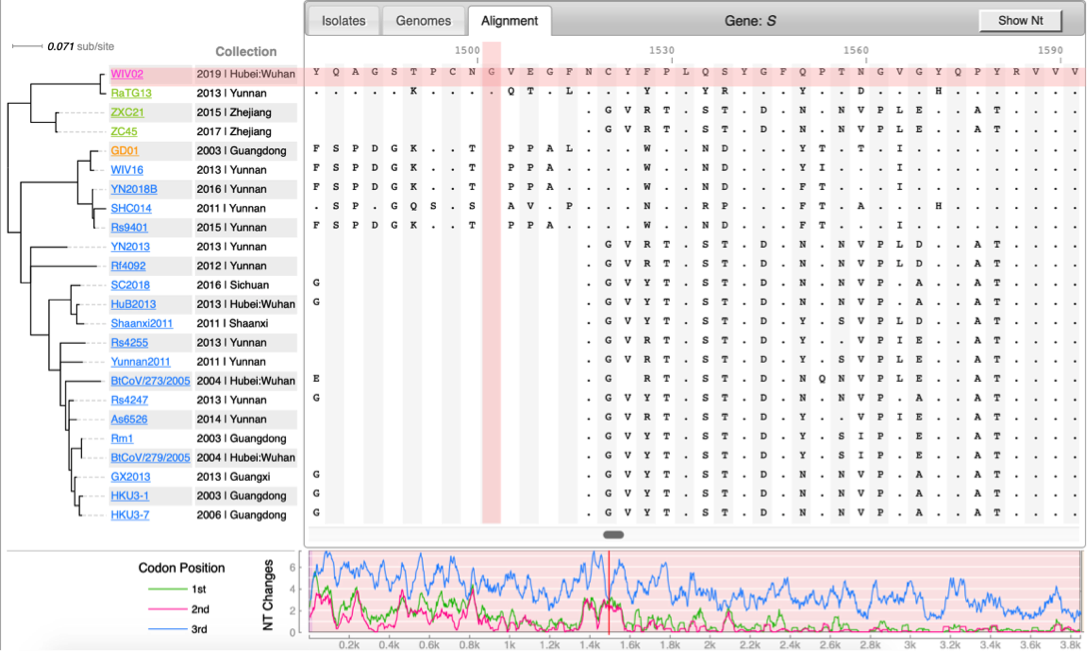
* [http://cov.genometracker.org Covid-19 Genome Tracker] | * [http://cov.genometracker.org Covid-19 Genome Tracker] | ||
== | ==CoV genome data set== | ||
* | * N=565 SARS-CoV-2 genomes collected during January & February 2020. Data source & acknowledgement [http://gisaid.org GIDAID] (<em>Warning: You need to acknowledge GISAID if you reuse the data in any publication</em>) | ||
* | * Download file: [http://diverge.hunter.cuny.edu/~weigang/qiu-akther.tar.gz data file] | ||
* | * Create a directory, unzip, & un-tar | ||
<syntaxhighlight lang='bash'> | |||
mkdir QiuAkther | |||
mv cov-camp.tar.gz QiuAkther/ | |||
cd QiuAkther | |||
tar -tzf cov-camp.tar.gz # view files | |||
tar -xzf cov-camp.tar.gz # un-zip & un-tar | |||
</syntaxhighlight> | |||
* View files | |||
<syntaxhighlight lang='bash'> | |||
file TCS.jar | |||
ls -lrt # long list, in reverse timeline | |||
less Jan-Feb.mafft # an alignment of 565 CoV2 genomes in FASTA format; "q" to quit | |||
less cov-565strains-617snvs.phy # non-gapped SNV alignment in PHYLIP format | |||
wc hap.txt # geographic origins | |||
head hap.txt | |||
wc group.txt # color assignment | |||
cat group.txt | |||
less cov-565strains.gml # graph file (output) | |||
</syntaxhighlight> | |||
==Learning Goals== | ==Bioinformatics Tools & Learning Goals== | ||
* BpWrapper: commandline tools for sequence, alignment, and tree manipulations (based on BioPerl). [https://github.com/bioperl/p5-bpwrapper Github Link] | * BpWrapper: commandline tools for sequence, alignment, and tree manipulations (based on BioPerl). | ||
* Haplotype network | ** [https://github.com/bioperl/p5-bpwrapper Github Link] | ||
** [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-018-2074-9/figures/1 Flowchart from publication] | |||
* Haplotype network with TCS [https://pubmed.ncbi.nlm.nih.gov/11050560/ PubMed link] | |||
* Web-interactive visualization with [http://D3js.org D3js] | |||
** [https://github.com/sairum/tcsBU Github link] | |||
** [https://cibio.up.pt/software/tcsBU/index.html Web tool] | |||
** [https://academic.oup.com/bioinformatics/article/32/4/627/1744448 Paper] | |||
==Tutorial== | ==Tutorial== | ||
* 2-2:30: Introduction on pathogen phylogenomics | * 2-2:30: Introduction on pathogen phylogenomics | ||
* 2:30-2:45: | * 2:30-2:45: Demo: sequence manipulation with BpWrapper | ||
<syntaxhighlight lang='bash'> | |||
bioseq --man | |||
bioseq -n Jan-Feb.mafft | |||
bioaln --man | |||
bioaln -n -i'fasta' Jan-Feb.mafft | |||
bioaln -l -i'fasta' Jan-Feb.mafft | |||
bioaln -n -i'phylip' cov-565strains-617snvs.phy | |||
bioaln -l -i'phylip' cov-565strains-617snvs.phy | |||
FastTree -nt cov-565strains-617snvs.phy > cov.dnd | |||
biotree --man | |||
biotree -n cov.dnd | |||
biotree -l cov.dnd | |||
<syntaxhighlight> | |||
* 2:45-3:00: build haplotype network with TCS | * 2:45-3:00: build haplotype network with TCS | ||
<syntaxhighlight lang='bash'> | |||
java -jar -Xmx1g TCS.jar | |||
<syntaxhighlight> | |||
* 3:00-3:15: interactive visualization with BuTCS | * 3:00-3:15: interactive visualization with BuTCS | ||
* 3:15-3:30: Q & A | * 3:15-3:30: Q & A | ||
Revision as of 07:23, 26 July 2020
| Lyme Disease (Borreliella) | CoV Genome Tracker | Coronavirus evolutuon |
|---|---|---|
Case studies from Qiu Lab
CoV genome data set
- N=565 SARS-CoV-2 genomes collected during January & February 2020. Data source & acknowledgement GIDAID (Warning: You need to acknowledge GISAID if you reuse the data in any publication)
- Download file: data file
- Create a directory, unzip, & un-tar
mkdir QiuAkther
mv cov-camp.tar.gz QiuAkther/
cd QiuAkther
tar -tzf cov-camp.tar.gz # view files
tar -xzf cov-camp.tar.gz # un-zip & un-tar
- View files
file TCS.jar
ls -lrt # long list, in reverse timeline
less Jan-Feb.mafft # an alignment of 565 CoV2 genomes in FASTA format; "q" to quit
less cov-565strains-617snvs.phy # non-gapped SNV alignment in PHYLIP format
wc hap.txt # geographic origins
head hap.txt
wc group.txt # color assignment
cat group.txt
less cov-565strains.gml # graph file (output)
Bioinformatics Tools & Learning Goals
- BpWrapper: commandline tools for sequence, alignment, and tree manipulations (based on BioPerl).
- Haplotype network with TCS PubMed link
- Web-interactive visualization with D3js
Tutorial
- 2-2:30: Introduction on pathogen phylogenomics
- 2:30-2:45: Demo: sequence manipulation with BpWrapper
<syntaxhighlight lang='bash'> bioseq --man bioseq -n Jan-Feb.mafft bioaln --man bioaln -n -i'fasta' Jan-Feb.mafft bioaln -l -i'fasta' Jan-Feb.mafft bioaln -n -i'phylip' cov-565strains-617snvs.phy bioaln -l -i'phylip' cov-565strains-617snvs.phy FastTree -nt cov-565strains-617snvs.phy > cov.dnd biotree --man biotree -n cov.dnd biotree -l cov.dnd <syntaxhighlight>
- 2:45-3:00: build haplotype network with TCS
<syntaxhighlight lang='bash'> java -jar -Xmx1g TCS.jar <syntaxhighlight>
- 3:00-3:15: interactive visualization with BuTCS
- 3:15-3:30: Q & A