imported>Lab |
imported>Weigang |
| Line 1: |
Line 1: |
| <span style="color: Seagreen;font-weight:bold;font-size:large;">Lab 12. Bioinformatics Exercises: BLAST & Genomes | | <center>Bioinformatics Boot Camp for Ecology & Evolution: '''Pathogen Evolutionary Genomics'''</center> |
| ==Expected Learning Outcomes==
| | <center>Thursday, Aug 6, 2020, 2 - 3:30pm</center> |
| * Be able to perform NCBI BLAST search for homologous sequences in GenBank.
| | <center>'''Instructors:''' Dr Weigang Qiu & Ms Saymon Akther</center> |
| * Be able to identify homologs in other model organisms.
| | <center>'''Email:''' weigang@genectr.hunter.cuny.edu</center> |
| * Be able to identify alternative splice forms a single gene using NCBI web tools
| | <center>'''Lab Website:''' http://diverge.hunter.cuny.edu/labwiki/</center> |
| * Be able to analyze locus structure from the information obtained from locus page
| |
| ----
| |
| | |
| ==Lab Report III==
| |
| # The lab report is worth 50 points.
| |
| # You have to complete the lab report by using the MS Word lab report template provided on Blackboard.
| |
| # Make sure to name your file with your LAST NAME-Lab12.
| |
| # you need to EMAIL your file to your T.A. to get credit for your work.
| |
| | |
| ----
| |
| | |
| ==Introduction==
| |
| Research in molecular genetics requires effective use of online bioinformatic tools to analyze and understand the genetic materials being worked with. The following exercises will expose you to real-world scenarios and introduce you to the methods and tools you can use to solve these problems.
| |
| | |
| In biology, homology is defined as a common or shared evolutionary origin. Therefore, homologous sequences are sequences diverged from a common ancestor. Note that the word "homology" is different from "similarity": homologous structures or sequences may not be similar (e.g., forearms in mammals and birds) and, conversely, similar structures or sequences may not be homologous (e.g., wings in birds and bats).
| |
| | |
| BLAST is a computer algorithm allowing for efficient search of similar sequences in a large database. While BLAST performs a similar function to Google search, you should not use Google to look for similar sequences in a human or other genome. When sequences are similar with a sufficient statistical significance (measured by e-value, see below), we consider these sequences homologous to each other.
| |
| ----
| |
| ==Exercise 1. Homology searching using BLAST==
| |
| # Go to the NCBI-BLAST website at [http://blast.ncbi.nlm.nih.gov/Blast.cgi NCBI/BLAST Home Page]
| |
| # To know more about BLAST, read the expanded answer by clicking on "Learn more"
| |
| # Since BLAST finds matches between nucleotide or protein sequences, it needs a "query" sequence as input as well as a "database" to search against. Make sure to know what your "query" sequence is and find the appropriate "database".
| |
| # Start BLASTing against the mouse genome by clicking "Mouse" under "BLAST Genomes"
| |
| # Copy and paste the following sequence into the "Enter Query Sequence" box:
| |
| <div style="font-family:Monospace;line-height:1;width:550px;border-style:solid;border-width:1px;border-color:#AAAAFF;background-color:#EEEEFF;padding-left:5px;padding-right:5px;padding-top:0px;padding-bottom:0px;">
| |
| CTAGATGCATTTACGAAGGAGACAGAAAACGTCTTTCGGCAATAGCTCTCAAATGCAAAACGACGTCGG
| |
| CGAGCTGTCCCTTACCTGGAGGCCCGCAGGAGAAGCGCGGTGATCCGAGAGGGTCCCCCAGGGGTGTCCG
| |
| GTCGGTCTCCCGCTCGCCCAGCAGACGGCTGCGGAAACGGGGCAGCGTTTAAATAACCCCAGCTGGAGAC
| |
| ATGTCAGGACTTAGCTCCTCCGACAGCCGACGCCGGACGTGTCCCAACTTGACCAGCCCCACAGGAAGAG
| |
| CTGAGTCAACTCGGCCCAGCCCAGTCCCACCCGTCCCGGAAGCCGCATCCCGGCGAGTCCGGGACCAGGC
| |
| ACCTGTCACCTCCTGGACCCCAGCAACGAGCCCAGCGCGACCCCGGAGCGGGCCCGAATTCT
| |
| </div>
| |
| <ol start="6">
| |
| <li>Scroll down to the bottom of the page and click "BLAST"
| |
| <li>Wait for 10-30 seconds for the results to return ('''be patient'''). Once the result page is loaded, locate and copy/write down
| |
| the following information in your lab report file for the first hit:
| |
| <ol>
| |
| <li>Species and strain | |
| <li>Chromosome
| |
| <li>Length of your query sequence
| |
| <li>Percent identity, number of matched bases, and number of gaps between the matched sequences
| |
| </ol>
| |
| <li>Click "Genome Data Viewer" at top right will bring you to a genome browser
| |
| <li>Mouse-over the central segment and click the link "GenBank View". A standard GenBank file of this gene will load. Locate the 1st "mRNA" feature block and write down the following structural information about this gene in your lab report file:
| |
| <ol> | |
| <li>Gene ID | |
| <li>Total length of the gene
| |
| <li>Number of introns
| |
| <li>Which is the non-template (mRNA analog) strand: the above sequence itself or its reverse complement? [Hint: note the word '''complement''' in mRNA and cDNA lines)
| |
| </ol> | |
| </ol> | |
| ----
| |
| | |
| ==Exercise 2. Explore the structure of human ''mdm2'' gene==
| |
| # Search [http://www.ncbi.nlm.nih.gov/sites/entrez?db=Nucleotide GenBank] using the accession AF527840. Read the GenBank file and find out from the feature table how many introns and exons this sequence has according to the "mRNA" and "CDS" features.
| |
| # Click on "mRNA" and notice that exon sequences are now highlighted
| |
| # Fill in '''Table 1''' in your lab report file for each EXON you could identify:
| |
| # Fill in '''Table 2''' in your lab report file for each INTRON you could identify:
| |
| # Click on "CDS" and notice that coding sequences are now highlighted
| |
| # Fill in '''Table 3''' in your lab report file for each coding sequence you could identify:
| |
| # Obtain the intron/exon gene structure and copy into your lab report file. To do this:
| |
| ##go back to the Genbank page for AF527840 (as instructed above)
| |
| ##click on the "Graphics" link
| |
| ##you will see a window with a diagram, showing the genomic sequence in green, the primary transcript in purple, and the coding sequence in red
| |
| ##copy and paste this diagram into your lab report (or create a desktop picture, crop as needed and paste into your lab report file)
| |
| # Answer the following questions, in your lab report file:
| |
| ## What is the total length of exons, introns, and coding sequences of this gene?
| |
| ## Are all exon sequences code for proteins? Which exons are non-coding in mdm2?
| |
| ## Align the first 5 bases of all introns. Which bases are conserved near intron start ("donor site")?
| |
| ## Align the last 5 bases of all introns. Which bases are conserved near intron end ("acceptor site")?
| |
| ## Using [http://weblogo.berkeley.edu/ WebLogo] and make a sequence logo for the acceptor site and another sequence logo for the donor site. To do so, copy & paste individual sequences at the acceptor site into [http://weblogo.threeplusone.com/create.cgi this text box] and click "Create Logo". Save the resulting image file and paste it into your lab report file. Repeat for the donor-site sequences.
| |
| <center> | | <center> |
| Table 1. ''mdm2'' Exons
| |
| {| class="wikitable"
| |
| |-
| |
| ! Exon # !! Start Position !! End Position !! Length
| |
| |-
| |
| | #1 || 1971 || 2271 || 301
| |
| |-
| |
| | #2 || ? || ? || ?
| |
| |}
| |
|
| |
| Table 2. ''mdm2'' Introns
| |
| {| class="wikitable"
| |
| |-
| |
| ! Intro Number !! Start Position !! End Position !! Length !! First 5 bases !! Last 5 bases !! Phase*
| |
| |-
| |
| | #1 || 2272 || 2987 || 616 || GTACT || TGTAG || ?
| |
| |-
| |
| | #2 || ? || ? || ? || ? || ? || ?
| |
| |}
| |
|
| |
| * Introns have phases. Phase 0 introns sit between 2 codons, phase 1 intron sit between the 1st codon position and the 2nd codon position, and phase 3 introns sit between the 2nd and 3rd codon position. How would you find out the phase of an intron? [Hint, use Table 3 CDS positions below].
| |
|
| |
| Table 3. ''mdm2'' Coding Sequences (CDS)
| |
| {| class="wikitable" | | {| class="wikitable" |
| |- | | |- |
| ! CDS # !! Start Position !! End Position !! Length | | ! Lyme Disease (Borreliella) !! CoV Genome Tracker !! Coronavirus evolutuon |
| |-
| |
| | #1 || 2992 || 3072 || 81
| |
| |- | | |- |
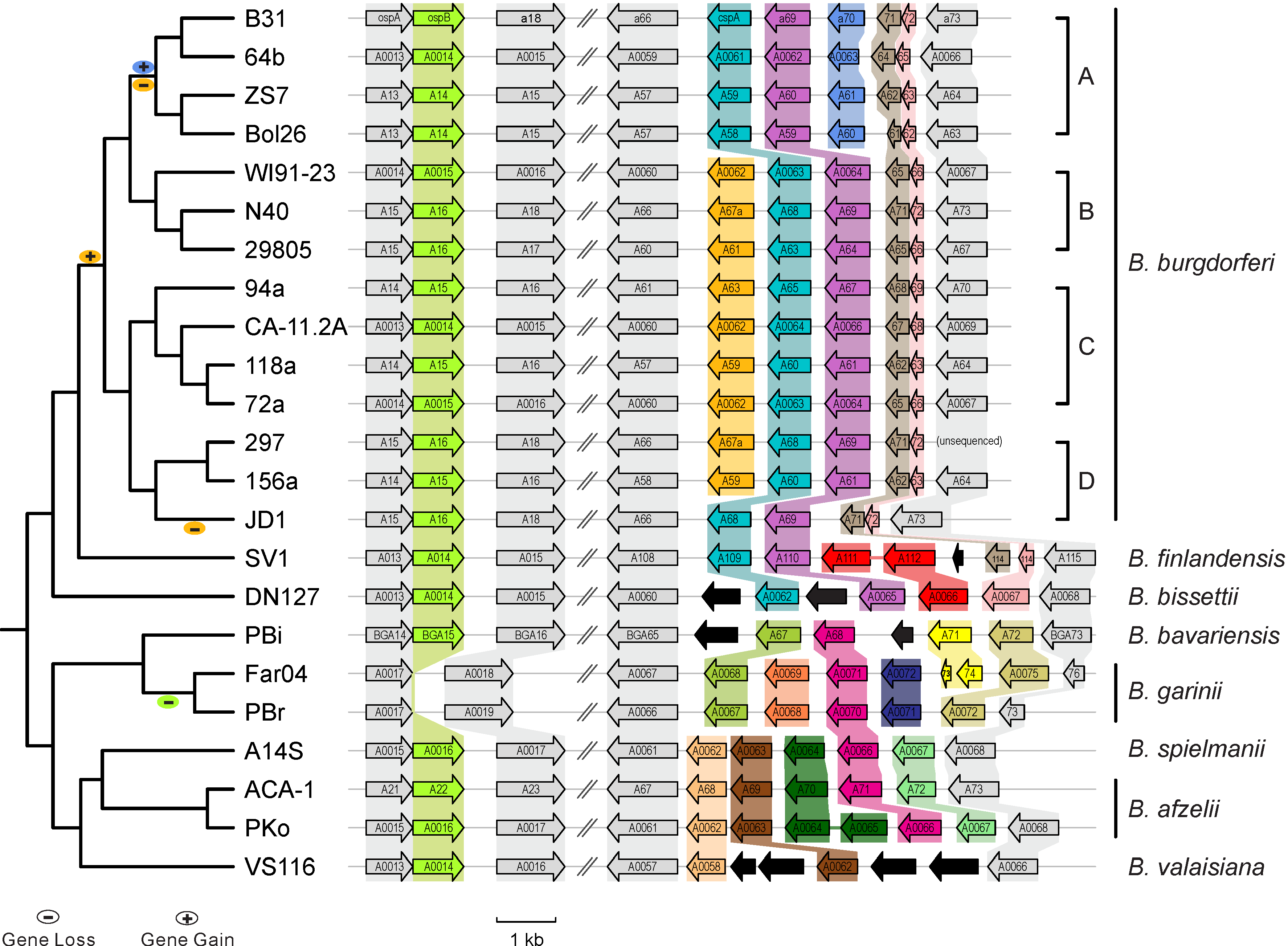
| | #2 || ? || ? || ? | | | [[File:Lp54-gain-loss.png|300px|thumbnail| Gains & losses of host-defense genes among Lyme pathogen genomes (Qiu & Martin 2014)]] || |
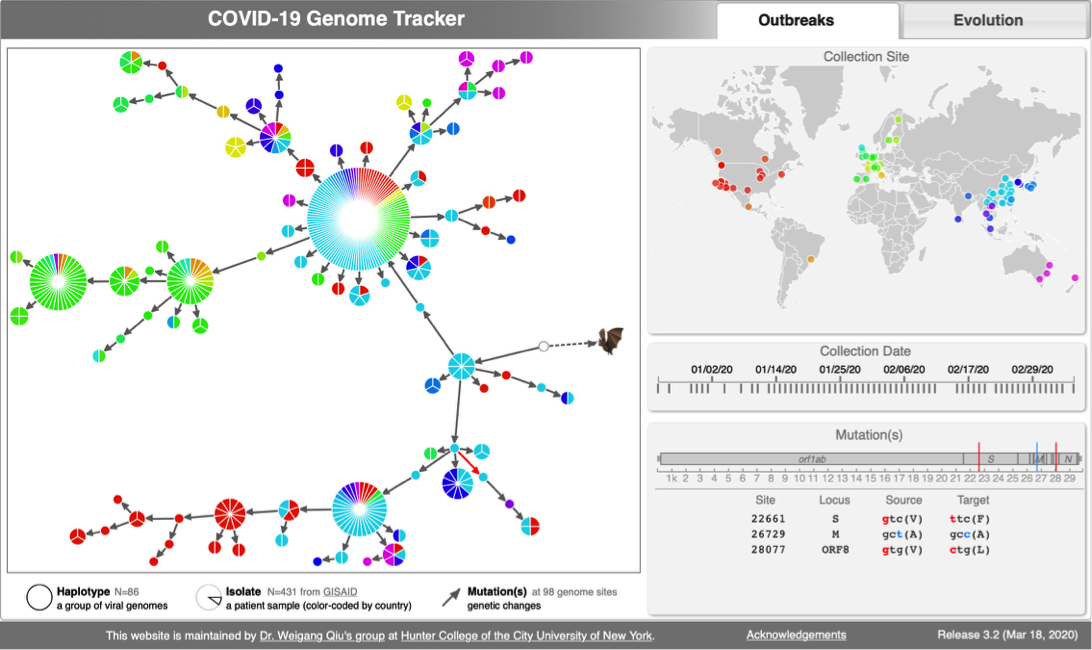
| | [[File:Cov-screenshot-1.png|300px|thumbnail| [http://cov.genometracker.org/ Haplotype network] ]] |
| | || |
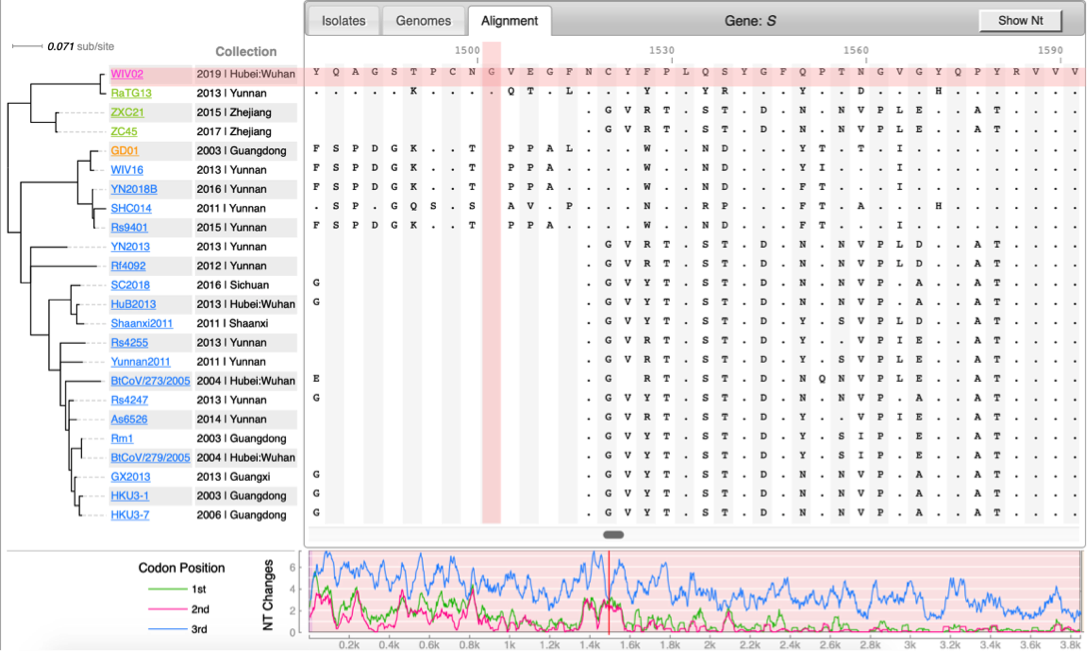
| | [[File:Cov-screenshot-2.png|300px|thumbnail| Spike protein alignment ]] |
| |} | | |} |
| </center> | | </center> |
| ---- | | ---- |
|
| |
|
| ==Exercise 3. MDM2 homologs in other species== | | ==Case studies from Qiu Lab== |
| This exercise will consist in comparing the predicted protein sequences of MDM2 in three species: human (H. sapiens), mouse (M. musculus) and zebrafish (D. rerio).
| | * [http://borreliabase.org Comparative genomics of worldwide Lyme disease pathogens] |
| You will need to download the human MDM2 sequence, find the mouse and fish homologs, and copy each sequence into a MS Word file using the following format:
| | * [http://cov.genometracker.org Covid-19 Genome Tracker] |
| | |
| Names (First, Last)
| |
| [Blank line]
| |
| >Human MDM2
| |
| --your amino acid sequence here--
| |
| >Mouse MDM2
| |
| --your amino acid sequence here--
| |
| >Zebrafish MDM2
| |
| --your amino acid sequence here--
| |
|
| |
|
| | ==CoV genome data set== |
| | * N=565 SARS-CoV-2 genomes collected during January & February 2020. Data source & acknowledgement [http://gisaid.org GIDAID] (<em>Warning: You need to acknowledge GISAID if you reuse the data in any publication</em>) |
| | * Download file: [http://diverge.hunter.cuny.edu/~weigang/qiu-akther.tar.gz data file] |
| | * Create a directory, unzip, & un-tar |
| | <syntaxhighlight lang='bash'> |
| | mkdir QiuAkther |
| | mv cov-camp.tar.gz QiuAkther/ |
| | cd QiuAkther |
| | tar -tzf cov-camp.tar.gz # view files |
| | tar -xzf cov-camp.tar.gz # un-zip & un-tar |
| | </syntaxhighlight> |
| | * View files |
| | <syntaxhighlight lang='bash'> |
| | file TCS.jar |
| | ls -lrt # long list, in reverse timeline |
| | less Jan-Feb.mafft # an alignment of 565 CoV2 genomes in FASTA format; "q" to quit |
| | less cov-565strains-617snvs.phy # non-gapped SNV alignment in PHYLIP format |
| | wc hap.txt # geographic origins |
| | head hap.txt |
| | wc group.txt # color assignment |
| | cat group.txt |
| | less cov-565strains.gml # graph file (output) |
| | </syntaxhighlight> |
|
| |
|
| | ==Bioinformatics Tools & Learning Goals== |
| | * BpWrapper: commandline tools for sequence, alignment, and tree manipulations (based on BioPerl). |
| | ** [https://github.com/bioperl/p5-bpwrapper Github Link] |
| | ** [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-018-2074-9/figures/1 Flowchart from publication] |
| | * Haplotype network with TCS [https://pubmed.ncbi.nlm.nih.gov/11050560/ PubMed link] |
| | * Web-interactive visualization with [http://D3js.org D3js] |
| | ** [https://github.com/sairum/tcsBU Github link] |
| | ** [https://cibio.up.pt/software/tcsBU/index.html Web tool] |
| | ** [https://academic.oup.com/bioinformatics/article/32/4/627/1744448 Paper] |
|
| |
|
| First, you will download the human MDM2 protein sequence. You will then use this sequence as a query to identify the mouse and zebrafish sequences. Follow these steps:
| | ==Tutorial== |
| | | * 2-2:30: Introduction on pathogen phylogenomics |
| # Go to this link: https://www.ncbi.nlm.nih.gov/genome/guide/human/
| | * 2:30-2:45: Demo: sequence manipulation with BpWrapper |
| # in the box "search for human genes" type in "MDM2"
| | <syntaxhighlight lang='bash'> |
| # you will see many hits- the top one corresponds to the human MDM2 locus- click on the link
| | bioseq --man |
| # This is the human MDM2 locus page- there is much information here. Scroll down to "Genomic regions, transcripts, and products"
| | bioseq -n Jan-Feb.mafft |
| # You see here a map of the known transcripts produced for this locus
| | bioaln --man |
| # now scroll down to "mRNA and Protein(s)"
| | bioaln -n -i'fasta' Jan-Feb.mafft |
| # here, find the entry corresponding to the LONGEST isoform
| | bioaln -l -i'fasta' Jan-Feb.mafft |
| # for each entry, you will see two identifiers : NM_.... and NP_....
| | bioaln -n -i'phylip' cov-565strains-617snvs.phy |
| # NM_... corresponds to the mRNA sequence for this isoform, and NP_.... to its predicted protein sequence
| | bioaln -l -i'phylip' cov-565strains-617snvs.phy |
| # click on the link for the protein sequence for the longest isoform, and find the 'FASTA' format
| | FastTree -nt cov-565strains-617snvs.phy > cov.dnd |
| # copy the protein sequence by highlighting all residues from the initial 'M' to the last residue- nothing else
| | biotree --man |
| # paste the sequence into your word file as instructed above
| | biotree -n cov.dnd |
| | | biotree -l cov.dnd |
| now let's find the mouse homolog using the human sequence as a query:
| | <syntaxhighlight> |
| | | * 2:45-3:00: build haplotype network with TCS |
| #go to the main NCBI link: https://www.ncbi.nlm.nih.gov
| | <syntaxhighlight lang='bash'> |
| #on the right side, under "Popular resources", click on "Blast"
| | java -jar -Xmx1g TCS.jar |
| #click on 'mouse' to blast the mouse genome- make sure you use the right tool (blastp) and the correct database (refuses protein)
| | <syntaxhighlight> |
| #in the window, paste in your human MDM2 sequence- this is your query, and click on "BLAST"
| | * 3:00-3:15: interactive visualization with BuTCS |
| #wait a few minutes... you will see your screen refreshing a few times
| | * 3:15-3:30: Q & A |
| #you get a number of hits- scroll down to the best one (under "alignments") and click on "gene" on the right side, under "related information"
| |
| #you are now on the mouse MDM2 locus page: find the protein sequence to the LONGEST isoform and paste into your page as above
| |
| | |
| now let us get the Zebrafish homolog:
| |
| | |
| #go to the genome portal : http://zfin.org
| |
| #find the protein sequence and paste into your page as above. Make sure you use the right program and database for a protein BLAST!
| |
| | |
| you will now make an alignment of all three sequences to see potential identities or similarities between them
| |
| | |
| #this involves two steps: first, the production of an output file by a program called "Clustal W"
| |
| # and second, the processing of this file to generate an alignment figure by a program called "Boxshade"
| |
| #go to this link: http://www.genome.jp/tools-bin/clustalw
| |
| #in the top window, paste in your three sequence by selecting from your first ">" sign to the end of your file (do not take your header, with your names)
| |
| # click on "multiple alignment"
| |
| #you will see an 'aln' output file: select the file including the header on top "CLUSTAL 2.1 multiple sequence alignment" down to the bottom of the file (no extra spaces)- copy in the buffer
| |
| #to do the alignment figure, go to : https://embnet.vital-it.ch/software/BOX_form.html
| |
| #there, enter your input sequence format as 'ALN' and in the window below that, paste your aln file
| |
| #click on 'run boxshade'
| |
| #under results: click on 'boxshade output 1"--- here's your alignment! (this should open with adobe acrobat and might take a bit of time)
| |
| #the output is a pdf file-- save it and import into your lab report word file (as page 2) by doing an "Insert--- picture from file" in MS Word- you will have two pages: page 1 with your MDM2 sequences, and page 2 with your alignment
| |
| | |
| | |
| | |
| | |
| ==Additonal questions: answer 3 questions from the ones shown below and include in your lab report--==
| |
| # Explain the following BLAST terms: “Expect” (e-value) [http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=FAQ#expect Read this FAQ], “Identities”, “Gap”, “Strand”.
| |
| # Which is a statistically more significant match by BLAST, a match with an e-value=1e-5 or a match with an e-value of 1?
| |
| # List and describe individual elements of a typical human gene based on mdm2.
| |
| #what are two determinants that can lead to the production of isoforms for a specific locus?
| |
| # What is the "GT-AG" rule? Explain how to read the sequence logos. Explain the significance of sequence conservation at exon-intron junctions.
| |
| # Discuss biological significance of alternative splicing, using mdm2 gene as an example.
| |
| #look at your alignment from part III: what are the black boxes- the grey boxes?
| |
| #do you see many gaps/insertions?do you think there is a pattern?
| |
| | |
| ---- | |