imported>Weigang |
imported>Weigang |
| Line 1: |
Line 1: |
| <center>'''Biomedical Genomics'''</center> | | <center>Bioinformatics Boot Camp for Ecology & Evolution: '''Pathogen Evolutionary Genomics'''</center> |
| <center>July 8-19, 2019</center> | | <center>Thursday, Aug 6, 2020, 2 - 3:30pm</center> |
| <center>'''Instructor:''' Weigang Qiu, Ph.D.<br>Professor, Department of Biological Sciences, City University of New York, Hunter College & Graduate Center<br>Adjunct Faculty, Department of Physiology and Biophysics, | | <center>'''Instructors:''' Dr Weigang Qiu & Ms Saymon Akther</center> |
| Institute for Computational Biomedicine, Weil Cornell Medical College</center>
| |
| <center>'''Office:''' B402 Belfer Research Building, 413 East 69th Street, New York, NY 10021, USA</center>
| |
| <center>'''Email:''' weigang@genectr.hunter.cuny.edu</center> | | <center>'''Email:''' weigang@genectr.hunter.cuny.edu</center> |
| <center>'''Lab Website:''' http://diverge.hunter.cuny.edu/labwiki/</center> | | <center>'''Lab Website:''' http://diverge.hunter.cuny.edu/labwiki/</center> |
| <br> | | <center> |
| <center>'''Host''': Shunqin Zhu (祝顺琴), Ph.D.<br>Associate Professor, School of Life Science, South West University</center>
| |
| ----
| |
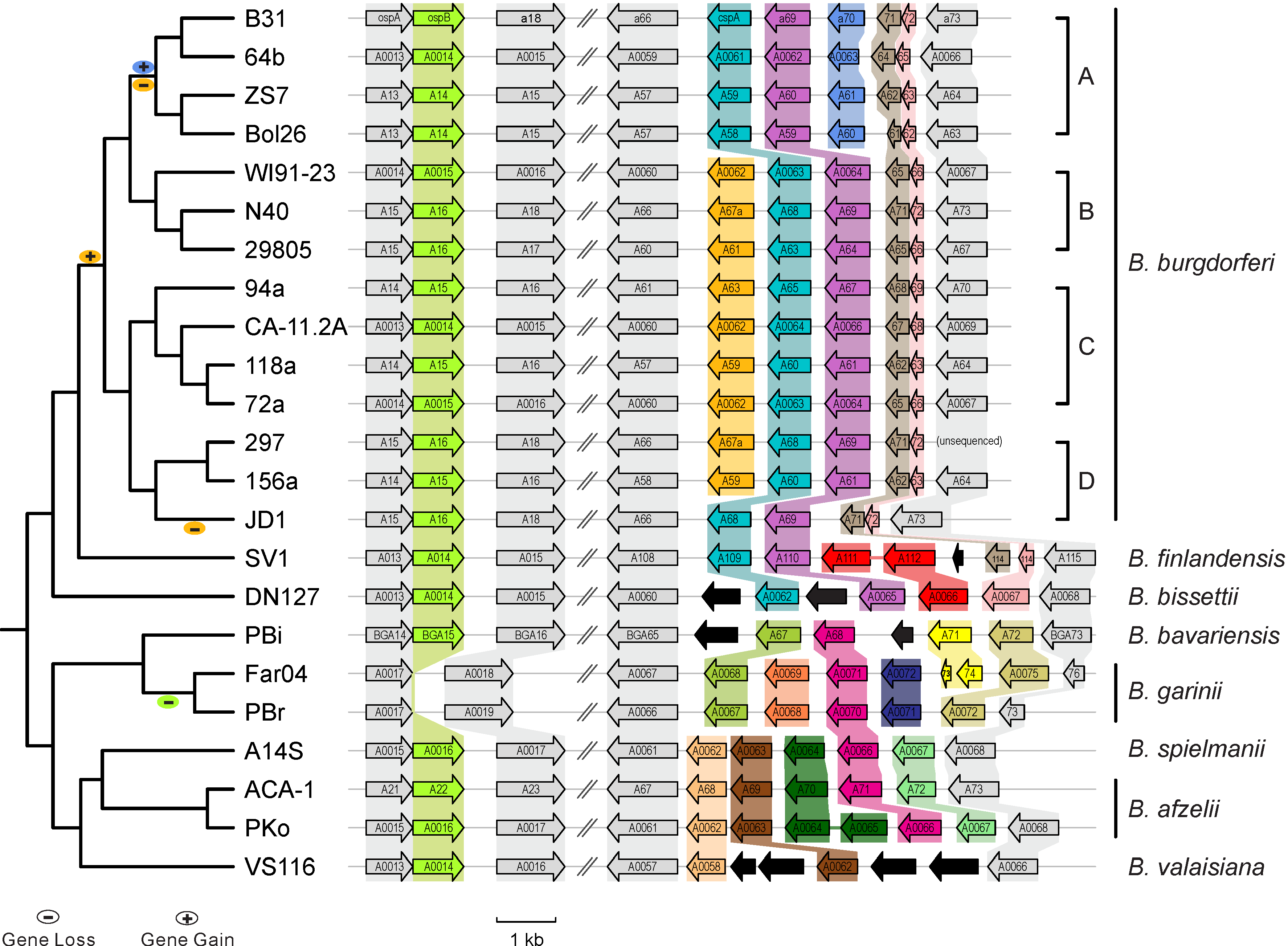
| [[File:Lp54-gain-loss.png|300px|thumbnail|Figure 1. Gains & losses of host-defense genes among Lyme pathogen genomes ([https://www.ncbi.nlm.nih.gov/pubmed/24704760 Qiu & Martin 2014])]]
| |
| ==Course Overview==
| |
| Welcome to BioMedical Genomics, a computer workshop for advanced undergraduates and graduate students. A genome is the total genetic content of an organism. Driven by breakthroughs such as the decoding of the first human genome and next-generation DNA -sequencing technologies, biomedical sciences are undergoing a rapid and irreversible transformation into a highly data-intensive field.
| |
| | |
| Genome information is revolutionizing virtually all aspects of life sciences including basic research, medicine, and agriculture. Meanwhile, use of genomic data requires life scientists to be familiar with concepts and skills in biology, computer science, as well as data analysis.
| |
| | |
| This workshop is designed to introduce computational analysis of genomic data through hands-on computational exercises, using published studies.
| |
| | |
| The pre-requisites of the course are college-level courses in molecular biology, cell biology, and genetics. Introductory courses in computer programming and statistics are preferred but not strictly required.
| |
| | |
| ==Learning goals==
| |
| By the end of this course successful students will be able to:
| |
| * Describe next-generation sequencing (NGS) technologies & contrast it with traditional Sanger sequencing
| |
| * Explain applications of NGS technology including pathogen genomics, cancer genomics, human genomic variation, transcriptomics, meta-genomics, epi-genomics, and microbiome.
| |
| * Visualize and explore genomics data using RStudio
| |
| * Replicate key results using a raw data set produced by a primary research paper
| |
| | |
| ==Web Links==
| |
| * Install R base: https://cloud.r-project.org
| |
| * Install R Studio (Desktop version): http://www.rstudio.com/download
| |
| * Download: [http://www.r4all.org/books/datasets R datasets]
| |
| * A reference book: [https://r4ds.had.co.nz/ R for Data Science (Wickharm & Grolemund)]
| |
| | |
| ==Quizzes and Exams==
| |
| Student performance will be evaluated by attendance, three (4) quizzes and a final report:
| |
| * Attendance: 50 pts
| |
| * Assignments: 5 x 10 = 50 pts
| |
| * Open-book Quizzes: 2 x 25 pts = 50 pts
| |
| * Take-home Mid-term: 50 pts
| |
| * Final presentation: 50 pts
| |
| Total: 250 pts
| |
| | |
| ==Course Schedule==
| |
| {| class="wikitable" | | {| class="wikitable" |
| |- | | |- |
| ! Date & Hour !! Tutorials !! Assignment !! Quiz & Exam | | ! Lyme Disease (Borreliella) !! CoV Genome Tracker !! Coronavirus evolutuon |
| |- | | |- |
| | July 8 (Mon), 8:40-12:10 || Introduction; R Tutorial I; | | | [[File:Lp54-gain-loss.png|300px|thumbnail| Gains & losses of host-defense genes among Lyme pathogen genomes (Qiu & Martin 2014)]] || |
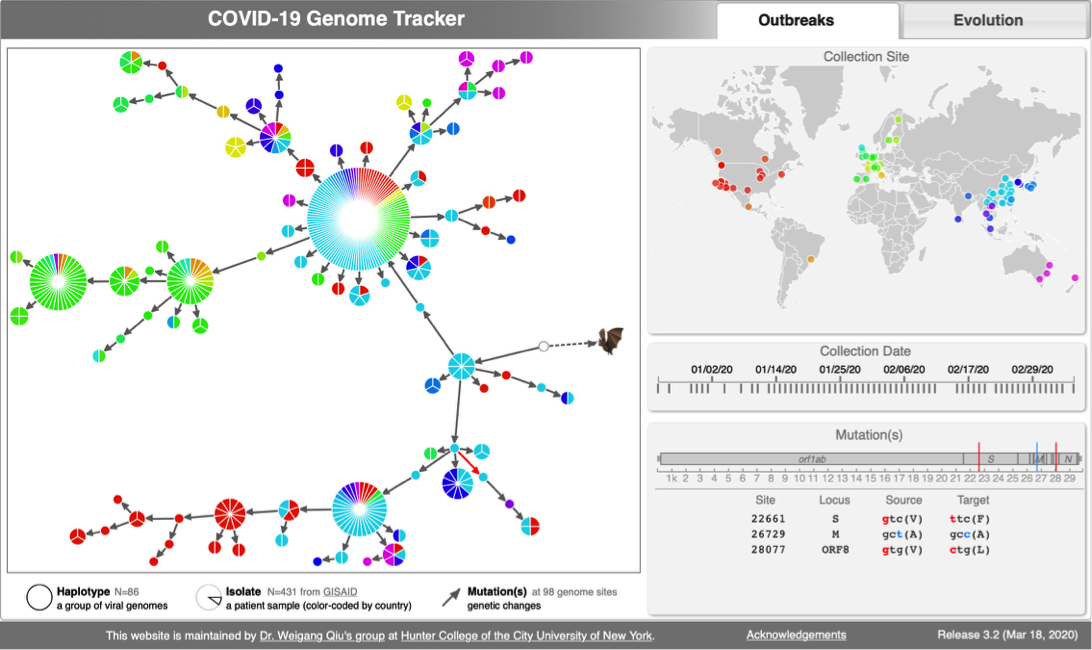
| [[File:R-part-1-small.pdf|thumbnail|Lecture slides]] | | [[File:Cov-screenshot-1.png|300px|thumbnail| [http://cov.genometracker.org/ Haplotype network] ]] |
| ||
| |
| Assignment #1 (create a WORD document including scripts & graphs (i.e., compile your work into a lab report, due tomorrow)
| |
| * Install R/R studio and the "tidyverse" package on your own computer
| |
| * Recreate Script 1 & Mini-Practical
| |
| * Show help page for function "seq"
| |
| * Download dataset
| |
| ** Create a new folder (e.g., Desktop/rtutor)
| |
| ** Create a sub-folder (e.g., Desktop/rtutor/data/)
| |
| ** Download from http://www.r4all.org/the-book/datasets
| |
| ** Save to the sub-folder
| |
| ** Unzip the file
| |
| ||
| |
| |- | |
| | July 9 (Tu), 8:40-12:10 || R Tutorials II && III,
| |
| [[File:R-part-2.pdf|thumbnail|Lecture slides]] | |
| ||
| |
| Assignment #2
| |
| * The following is a portion of the dataset of Mycobacterium growth (kindly shared by Aswad from Dr Xie's lab). It shows OD (optical density) values. Transform this table ("wide" format) into the "tall/tidy" format (use paper & pen, no need to use R studio or any computer program):
| |
| {| class="wikitable"
| |
| |-
| |
| ! Hour !! Control !! Gene !! Control.with.Arg !! Gene.with.Arg
| |
| |-
| |
| | 0 || 0.06 || 0.022 || 0.031 || 0.01
| |
| |-
| |
| | 4 || 0.087 || 0.102 || 0.082 || 0.081
| |
| |-
| |
| | 8 || 0.113 || 0.185 || 0.086 || 0.135
| |
| |}
| |
| * In R studio, read the dataset from the file "FlowerColourVisits.csv" and save it into an object named as "flower"
| |
| ** Show head, tail, dimension of the data frame "flower"
| |
| ** Show data summary with "summary" & "glimpse" commands. Which column is a categorical data type?
| |
| ** Select the column named "colour"
| |
| ** Select rows from the 3rd to the 20th
| |
| ** Select the 3rd, 10th, and 20th rows
| |
| ** Select only the rows that have the colour of "red" (hint use <code>colour=="red"</code>
| |
| ** Create a new column, named "logVisit", that is log(1+number.of.visit)
| |
| ** Sort the "flower" data by the column "number.of.visit"
| |
| ** Perform the following data transformation using the chaining operator (i.e., "%>%"): Select rows from the 3rd to the 20th, then filter by colour of "red", and then show head
| |
| ** Obtain the mean number of visit for each colour as a group (Hint: use "group_by" & "summarise")
| |
| ||
| |
| |-
| |
| | July 10 (Wed), 8:40-12:10 || R Tutorial IV
| |
| [[File:R-part-3.pdf|thumbnail|Lecture slides]]
| |
| || | | || |
| Assignment #3
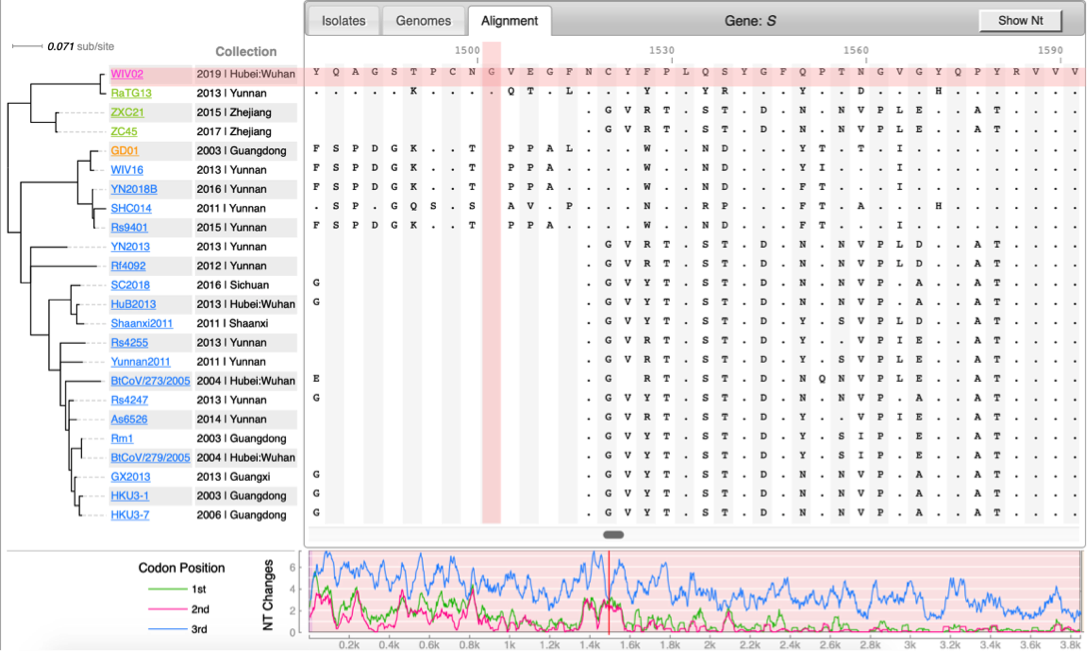
| | [[File:Cov-screenshot-2.png|300px|thumbnail| Spike protein alignment ]] |
| {| class="wikitable"
| |
| |-
| |
| ! Task!! Graph
| |
| |-
| |
| | Use the "iris" dataset to reproduce the plot shown at right (Hint: load data with <code>data(iris)</code>) ||
| |
| [[File:Iris-1.png|200px|thumbnail]] | |
| |-
| |
| | Use the "flower" dataset (see Assignment #2 on how to load data) to reproduce the plot shown at right ||
| |
| [[File:Flower-1.png|200px|thumbnail]]
| |
| |} | | |} |
| | </center> |
| | ---- |
|
| |
|
| || Quiz I
| | ==Case studies from Qiu Lab== |
| |-
| | * [http://borreliabase.org Comparative genomics of worldwide Lyme disease pathogens] |
| | July 11 (Thur), 8:40-12:10 || Intro to NGS; R Tutorial V ||
| | * [http://cov.genometracker.org Covid-19 Genome Tracker] |
| | |
| | ==CoV genome data set== |
| | * N=565 SARS-CoV-2 genomes collected during January & February 2020. Data source & acknowledgement [http://gisaid.org GIDAID] (<em>Warning: You need to acknowledge GISAID if you reuse the data in any publication</em>) |
| | * Download file: [http://diverge.hunter.cuny.edu/~weigang/qiu-akther.tar.gz data file] |
| | * Create a directory, unzip, & un-tar |
| | <syntaxhighlight lang='bash'> |
| | mkdir QiuAkther |
| | mv cov-camp.tar.gz QiuAkther/ |
| | cd QiuAkther |
| | tar -tzf cov-camp.tar.gz # view files |
| | tar -xzf cov-camp.tar.gz # un-zip & un-tar |
| | </syntaxhighlight> |
| | * View files |
| | <syntaxhighlight lang='bash'> |
| | file TCS.jar |
| | ls -lrt # long list, in reverse timeline |
| | less Jan-Feb.mafft # an alignment of 565 CoV2 genomes in FASTA format; "q" to quit |
| | less cov-565strains-617snvs.phy # non-gapped SNV alignment in PHYLIP format |
| | wc hap.txt # geographic origins |
| | head hap.txt |
| | wc group.txt # color assignment |
| | cat group.txt |
| | less cov-565strains.gml # graph file (output) |
| | </syntaxhighlight> |
|
| |
|
| ||
| | ==Bioinformatics Tools & Learning Goals== |
| Take-home mid-term (50 pts)
| | * BpWrapper: commandline tools for sequence, alignment, and tree manipulations (based on BioPerl). |
| * List pros & cons of Sanger vs NGS
| | ** [https://github.com/bioperl/p5-bpwrapper Github Link] |
| * Compare accuracy, read length, and error rate between Illumina and PacBio | | ** [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-018-2074-9/figures/1 Flowchart from publication] |
| * Describe sequence information captured with each of the following file formats: FASTA, FASTQ, SAM, VCF | | * Haplotype network with TCS [https://pubmed.ncbi.nlm.nih.gov/11050560/ PubMed link] |
| * Run t-test & regression analysis | | * Web-interactive visualization with [http://D3js.org D3js] |
| |-
| | ** [https://github.com/sairum/tcsBU Github link] |
| | July 12 - 14 (Fri, Sat & Sun) || (Weekend break; No class) || ||
| | ** [https://cibio.up.pt/software/tcsBU/index.html Web tool] |
| |-
| | ** [https://academic.oup.com/bioinformatics/article/32/4/627/1744448 Paper] |
| | July 15 (Mon), 8:00-12:10 || Case Study 1. Fish microbiome || Assignment #4
| |
| ||
| |
| |-
| |
| | July 16 (Tu), 8:00-12:10 || Case Study 2. Transcriptome || Assignment #5
| |
| ||
| |
| |-
| |
| | July 17 (Wed), 8:00-12:10 || Case Study 3. Lyme Disease || || Quiz II
| |
| |-
| |
| | July 18 (Thur), 8:00-12:10|| || || Presentations
| |
| |}
| |
|
| |
|
| ==Papers & Datasets== | | ==Tutorial== |
| {| class="wikitable sortable"
| | * 2-2:30: Introduction on pathogen phylogenomics |
| |-
| | * 2:30-2:45: Demo: sequence manipulation with BpWrapper |
| ! Omics Application !! Paper link !! Data set !! NGS Technology
| | <syntaxhighlight lang='bash'> |
| |-
| | bioseq --man |
| | Microbiome || [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0193652 Rimoldi_etal_2018_PlosOne] || [https://doi.org/10.1371/journal.pone.0193652.s004 S1 Dataset] || 16S rDNA amplicon sequencing
| | bioseq -n Jan-Feb.mafft |
| |-
| | bioaln --man |
| | Transcriptome || [https://science.sciencemag.org/content/350/6264/1096 Wang_etal_2015_Science] || Tables S2 & S4 || RNA-Seq
| | bioaln -n -i'fasta' Jan-Feb.mafft |
| |-
| | bioaln -l -i'fasta' Jan-Feb.mafft |
| | Transcriptome & Regulome || [https://bmcmedgenomics.biomedcentral.com/articles/10.1186/s12920-019-0477-8 Nava_etal_2019_BMCGenomics] || Tables S2 & S3 || RNA-Seq & CHIP-Seq
| | bioaln -n -i'phylip' cov-565strains-617snvs.phy |
| |-
| | bioaln -l -i'phylip' cov-565strains-617snvs.phy |
| | Proteome || [https://www.ncbi.nlm.nih.gov/pubmed/28232952 Qiu_etal_2017_NPJ] || (to be posted) || SILAC
| | FastTree -nt cov-565strains-617snvs.phy > cov.dnd |
| |-
| | biotree --man |
| | Population genomics (Lyme) || [https://jcm.asm.org/content/56/11/e00940-18.long Di_etal_2018_JCM] || [https://github.com/weigangq/ocseq Data & R codes] || Amplicon sequencing (antigen locus)
| | biotree -n cov.dnd |
| |-
| | biotree -l cov.dnd |
| | Population genomics/GWAS (Human) || [https://science.sciencemag.org/content/351/6274/737.long Simonti_etal_2016_Science] || [https://science.sciencemag.org/highwire/filestream/673591/field_highwire_adjunct_files/1/aad2149-Simonti-SM.Table.S2.xlsx Table S2] || whole-genome sequencing (WGS); [http://www.internationalgenome.org/ 1000 Genome Project (IGSR)]
| | <syntaxhighlight> |
| |-
| | * 2:45-3:00: build haplotype network with TCS |
| | TB surveillance || [https://jcm.asm.org/content/53/7/2230 Brow_etal_2015] || [https://www.ebi.ac.uk/ena/data/view/PRJEB9206 Sequence Archives]|| Whole-genome sequencing (WGS)
| | <syntaxhighlight lang='bash'> |
| |-
| | java -jar -Xmx1g TCS.jar |
| | Example || Example || Example || Example
| | <syntaxhighlight> |
| |-
| | * 3:00-3:15: interactive visualization with BuTCS |
| | Example || Example || Example || Example
| | * 3:15-3:30: Q & A |
| |-
| |
| | Example || Example || Example || Example
| |
| |}
| |