imported>Weigang |
imported>Weigang |
| Line 8: |
Line 8: |
| ---- | | ---- |
| [[File:Lp54-gain-loss.png|200px|thumbnail|Figure 1. Gains & losses of host-defense genes among Lyme pathogen genomes (Qiu & Martin 2014)]] | | [[File:Lp54-gain-loss.png|200px|thumbnail|Figure 1. Gains & losses of host-defense genes among Lyme pathogen genomes (Qiu & Martin 2014)]] |
| [[File:Igv mdpa.png|200px|thumbnail|Figure 2. Development of drug resistance mutation in Pseudomonas in a single cancer patient: top (April) vs. bottom (July) (Un-published data)]]
| | ==Course Overview== |
| ==What is evolutionary genomics?== | | Welcome to Introductory BioMedical Genomics, a computer workshop for advanced undergraduates and graduate students. A genome is the total genetic content of an organism. Driven by breakthroughs such as the decoding of the first human genome and next-generation DNA -sequencing technologies, biomedical sciences are undergoing a rapid and profound transformation into a highly data-intensive field. Genome information is revolutionizing virtually all aspects of life sciences including basic basic, medicine, and agriculture. Meanwhile, use of genomic data requires life scientists to be familiar with concepts and skills in biology, computer science, as well as data analysis. |
| Genomes differ among individuals and species. Evolutionary genomics studies genome variability and genome changes using evolutionary principles. Typical applications include identification of human genome variations associated with diseases and identification of pathogen virulence genes.
| |
|
| |
|
| Genome changes are studied at two distinct levels: (1) within-species/within-population variations (e.g., human genetic variation), and (2) between-species divergence (e.g., human-mouse comparisons).
| | This workshop is designed to introduce computational analysis of genomic data through hands-on computational exercises. |
|
| |
|
| The key for analyzing genome variations within species is "population-thinking", the idea that there is no one individual genome that is standard, normal, or disease-free. | | The pre-requisites of the course includes college-level courses in molecular biology, cell biology, and genetics. Introductory courses in computer programming and statistics are preferred but not strictly required. |
|
| |
|
| The key for comparing genomes across species is "tree-thinking", the idea that evolution happens by diversification (like a branching tree), not by climbing a ladder. There is no such thing as "advanced" or "primitive" species. All living species have the exact same evolutionary distances/time of divergence since the origin of life.
| | ==Learning outcomes== |
| | | By the end of this course successful students will be able to: |
| ==Case studies from Qiu Lab== | | * Describe next-generation sequencing (NGS) technologies & contrast it with traditional Sanger sequencing |
| * Between-sepcies genome comparisons: Comparative genomics of worldwide Lyme disease pathogens. [http://borreliabase.org/ BorreliaBase] (Figure 1)
| | * Explain applications of NGS technology including pathogen genomics, cancer genomics, human genomic variation, transcriptomics, meta-genomics, epi-genomics, and microbiome, and single-cell genomics |
| * Within-population genome comparison: Genomic epidemiology of Group B Streptococcus: [http://diverge.hunter.cuny.edu/~weigang/gbs-browser/%20 Gene gains & losses associated with Group B Streptococcus virulence] | | * Visualize and explore genomics data using RStudio |
| * Within-host genome evolution: Evolution of multi-drug antibiotic-resistance Pseudomonas in cancer patients (Figure 2) | | * Ability to replicate results using a data set associated with a primary research paper |
| | |
| ==Bioinformatics workflow for comparative analysis of bacterial pathogen genomes==
| |
| * Pathogen isolation -> DNA extraction -> Library preparation -> High-through sequencing
| |
| * De novo genome assembly (canu; velvet; etc)
| |
| * Identify reference genome from NCBI database (kraken)
| |
| * Variant call (bwa; cortex_var; samtools mpileup)
| |
| * Infer genome phylogeny (muscle; reXML)
| |
| * Annotation (PATRIC)
| |
| * Custom genome browser (JavaScript; D3 library for interactive graphics)
| |
|
| |
| ==Essential bioinformatics skills==
| |
| * Linux command-line interface (e.g., BASH shell)
| |
| * Familiarity with a programming language (e.g., Python or Perl)
| |
| * Data visualization & statistical analysis (e.g., JavaScript; the R statistical computing environment)
| |
| | |
| ==Textbooks for genome evolution==
| |
| * Graur, 2016, Molecular and Genome Evolution, First Edition, Sinauer Associates, Inc. ISBN: 978-1-60535-469-9. [http://www.sinauer.com/molecular-and-genome-evolution.html Publisher's Website]
| |
| * Baum & Smith, 2013. Tree Thinking: an Introduction to Phylogenetic Biology, Roberts & Company Publishers, Inc. | |
| | |
| ==Learning Goals==
| |
| * Be able to compare evolutionary relationships using phylogenetic trees | |
| * Be able to use command-line tools for batch-processing of genome files
| |
| * Be able to perform genome-wide association analysis on the R platform
| |
| | |
| ==Schedule==
| |
| * 9:00 - 9:25: Introduction; [http://rstudio.org Install R & R Studio]; Download fasta file & save as "ospC-pep.fasta" : [[File:OspC-pep.txt|thumbnail]]
| |
| * 9:30 - 10:00: Unix Tutorial ([http://korflab.ucdavis.edu/Unix_and_Perl/current.html#part1 Part I. Unix Basics])
| |
| * 10:05 - 10:30: Unix Tutorial ([http://korflab.ucdavis.edu/Unix_and_Perl/current.html#part2 Part II Advanced Unix])
| |
| * 10:35 - 11:00: Tree-thinking Quizzes: Slides [[File:Big-data-phylogeny.pptx|thumbnail]] & Handouts [[File:Pretest.pdf|thumbnail]]
| |
| * 11:05 - 12: Demo: [[Mini-Tutorals#ospC_amplicon_identification|identification of genomic mutations associated with antibiotic resistance]]
| |
| ** Slides on background
| |
| ** Demo (on wallace lab server)
| |
| | |
| ==Exercises & Challenges==
| |
| * Finish Tree Thinking Quizzes
| |
| * Unix exercises:
| |
| ** count the number of sequences using "grep -v" or "wc"
| |
| ** display the first 5 lines of a file
| |
| ** display the last 5 lines of a file
| |
| ** change upper-cases to lower-cases
| |
| ** change "|" to "_"
| |
| ** replace strings
| |
Biomedical Genomics
July 8-19, 2019
Instructor: Weigang Qiu, Ph.D.
Professor, Department of Biological Sciences, City University of New York, Hunter College & Graduate Center
Adjunct Faculty, Department of Physiology and Biophysics
Institute for Computational Biomedicine, Weil Cornell Medical College
Office: B402 Belfer Research Building, 413 East 69th Street, New York, NY 10021, USA
Email: weigang@genectr.hunter.cuny.edu
Lab Website: http://diverge.hunter.cuny.edu/labwiki/
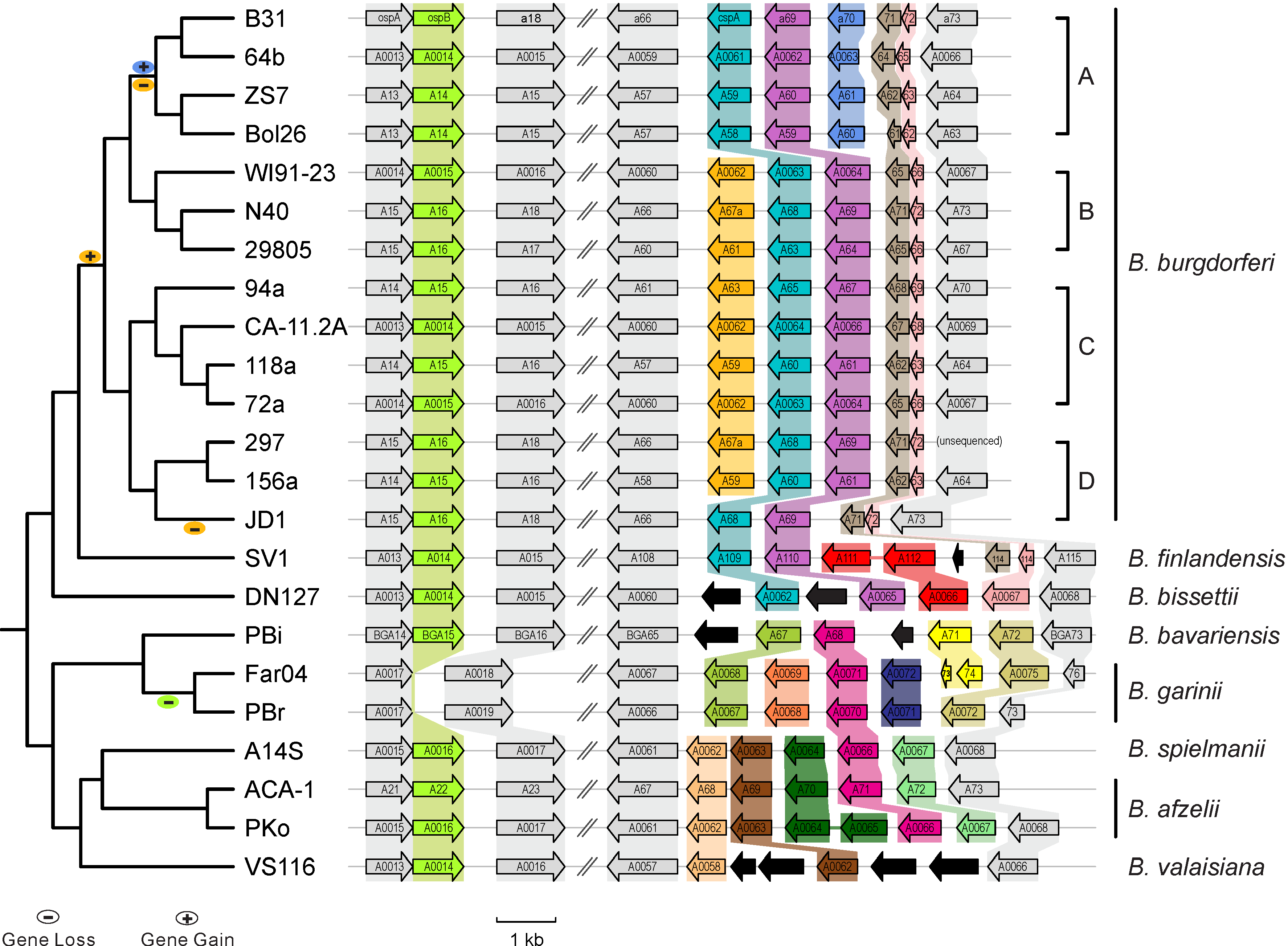
Figure 1. Gains & losses of host-defense genes among Lyme pathogen genomes (Qiu & Martin 2014)
Course Overview
Welcome to Introductory BioMedical Genomics, a computer workshop for advanced undergraduates and graduate students. A genome is the total genetic content of an organism. Driven by breakthroughs such as the decoding of the first human genome and next-generation DNA -sequencing technologies, biomedical sciences are undergoing a rapid and profound transformation into a highly data-intensive field. Genome information is revolutionizing virtually all aspects of life sciences including basic basic, medicine, and agriculture. Meanwhile, use of genomic data requires life scientists to be familiar with concepts and skills in biology, computer science, as well as data analysis.
This workshop is designed to introduce computational analysis of genomic data through hands-on computational exercises.
The pre-requisites of the course includes college-level courses in molecular biology, cell biology, and genetics. Introductory courses in computer programming and statistics are preferred but not strictly required.
Learning outcomes
By the end of this course successful students will be able to:
- Describe next-generation sequencing (NGS) technologies & contrast it with traditional Sanger sequencing
- Explain applications of NGS technology including pathogen genomics, cancer genomics, human genomic variation, transcriptomics, meta-genomics, epi-genomics, and microbiome, and single-cell genomics
- Visualize and explore genomics data using RStudio
- Ability to replicate results using a data set associated with a primary research paper
