EEB BootCamp 2020: Difference between revisions
Jump to navigation
Jump to search
Bioinformatics Boot Camp for Ecology & Evolution: Genomic Epidemiology
Thursday, Aug 6, 2020, 2 - 3:30pm
Instructors: Dr Weigang Qiu & Ms Saymon Akther
Email: weigang@genectr.hunter.cuny.edu
Lab Website: http://diverge.hunter.cuny.edu/labwiki/
imported>Weigang mNo edit summary |
imported>Lab |
||
| (97 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
<center>Bioinformatics Boot Camp for Ecology & Evolution: ''' | <center>Bioinformatics Boot Camp for Ecology & Evolution: '''Genomic Epidemiology'''</center> | ||
<center>Thursday, Aug 6, 2020, 2 - 3:30pm</center> | <center>Thursday, Aug 6, 2020, 2 - 3:30pm</center> | ||
<center>'''Instructors:''' Dr Weigang Qiu & Ms Saymon Akther</center> | <center>'''Instructors:''' Dr Weigang Qiu & Ms Saymon Akther</center> | ||
| Line 7: | Line 7: | ||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
! CoV Genome Tracker !! Coronavirus evolutuon !! Lyme Disease (Borreliella) | |||
|- | |- | ||
| | |||
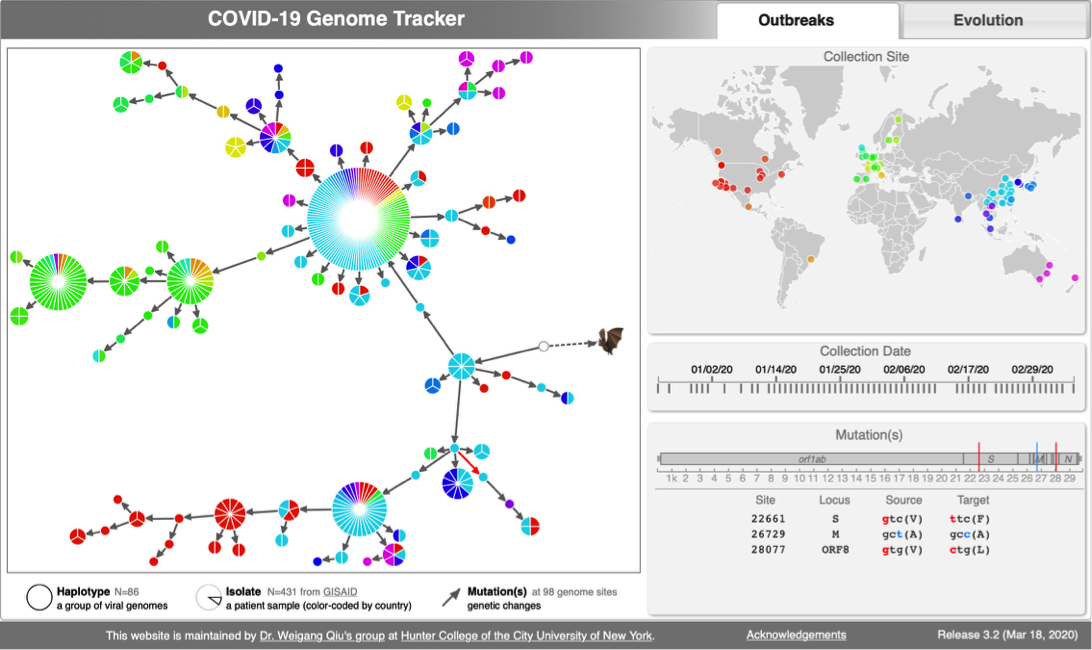
[[File:Cov-screenshot-1.png|300px|thumbnail| [http://cov.genometracker.org/ Haplotype network] ]] | [[File:Cov-screenshot-1.png|300px|thumbnail| [http://cov.genometracker.org/ Haplotype network] ]] | ||
|| | || | ||
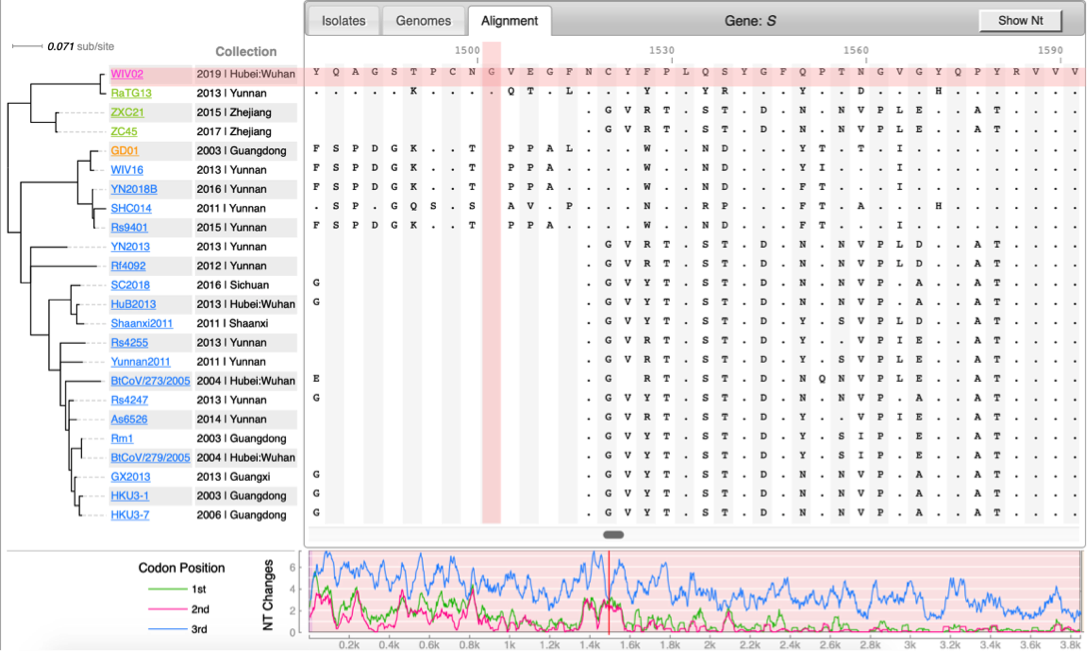
[[File:Cov-screenshot-2.png|300px|thumbnail| Spike protein alignment ]] | [[File:Cov-screenshot-2.png|300px|thumbnail| Spike protein alignment ]] | ||
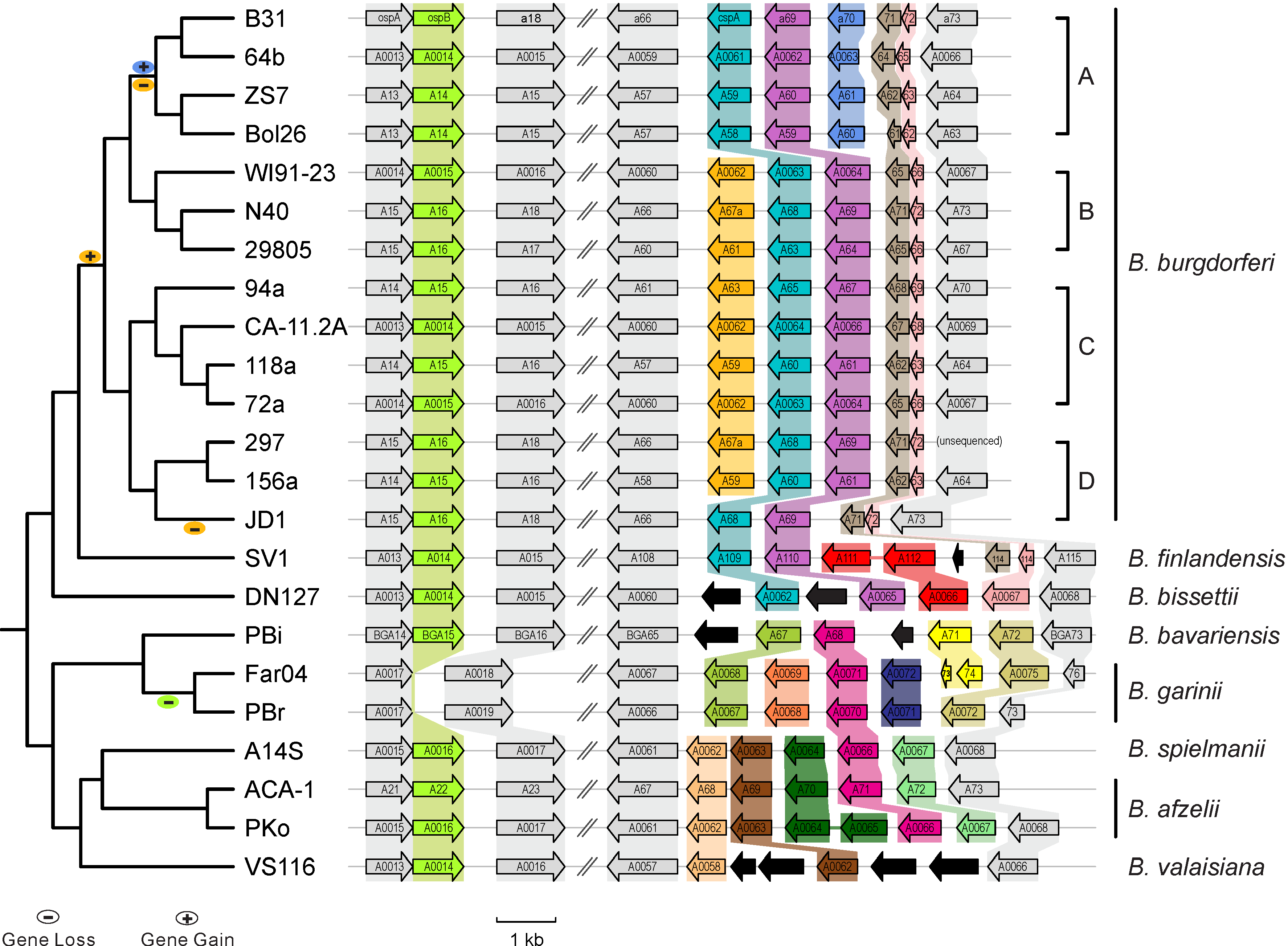
|| [[File:Lp54-gain-loss.png|300px|thumbnail| Gains & losses of host-defense genes among Lyme pathogen genomes (Qiu & Martin 2014)]] | |||
|} | |} | ||
</center> | </center> | ||
---- | ---- | ||
==Case studies | ==Case studies== | ||
* [http://cov.genometracker.org Covid-19 Genome Tracker] | |||
* [http://borreliabase.org Comparative genomics of worldwide Lyme disease pathogens] | * [http://borreliabase.org Comparative genomics of worldwide Lyme disease pathogens] | ||
* [http:// | * [https://nextstrain.org/ Next Strain] | ||
==Bioinformatics tools for genomic epidemiology== | |||
Required for the tutorial | |||
* bcftools: Reading/writing BCF2/VCF/gVCF files and calling/filtering/summarising SNP and short indel sequence variants [http://www.htslib.org/download/ Installation link] | |||
<syntaxhighlight lang='bash'> | |||
cd bcftools-1.10.2 | |||
./configure | |||
make | |||
make install | |||
## You may need sudo permissions to run make install | |||
bcftools --version | |||
# to check the man page | |||
man bcftools | |||
</syntaxhighlight> | |||
* vcftools: To work with genetic variation data in the form of VCF files [https://github.com/vcftools/vcftools Github link] [https://github.com/vcftools/vcftools/releases Build from Release Tarball] | |||
<syntaxhighlight lang='bash'> | |||
#download version v0.1.16 from second link | |||
cd vcftools-0.1.16 | |||
./configure | |||
make | |||
make install | |||
## You may need sudo permissions to run make install | |||
## Alternative: if you have homebrew in your computer | |||
brew install vcftools | |||
vcftools --version | |||
# to check the man page | |||
man vcftools | |||
</syntaxhighlight> | |||
* Sequence format converter [https://www.hiv.lanl.gov/content/sequence/FORMAT_CONVERSION/form.html Web tool] | |||
* TCS: To build Haplotype network, TCS.jar file is provided in the dataset folder below, Required Java. [https://pubmed.ncbi.nlm.nih.gov/11050560/ PubMed link] | |||
* Web-interactive visualization of Haplotype Network with tcsBU [https://cibio.up.pt/software/tcsBU/index.html Web tool]; [https://academic.oup.com/bioinformatics/article/32/4/627/1744448 Paper] | |||
* library(ggplot2) (R) | |||
Not required for the tutorial. Recommended | |||
* BpWrapper: command-line tools for manipulation of sequences, alignment, and tree (based on BioPerl). [https://github.com/bioperl/p5-bpwrapper Github Link]; [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-018-2074-9/figures/1 Flowchart from publication] | |||
* Pairwise genome alignment with MUMMER: [https://github.com/mummer4/mummer Github link] | |||
* Samtools: Reading/writing/editing/indexing/viewing SAM/BAM/CRAM format [http://www.htslib.org/download/ Installation link] | |||
==CoV genome data set== | |||
* N=100 SARS-CoV-2 genomes collected during January, February & March 2020. Data source & acknowledgement [http://gisaid.org GIDAID] (<em>Warning: You need to acknowledge GISAID if you reuse the data in any publication</em>) | |||
* Download the folder "bootcamp_august_6th_2020": [https://drive.google.com/file/d/1HBrHOJL3DIYrqzO0LIo7sdHvFbq4IZ3n/view?usp=sharing data file] | |||
* unzip the folder | |||
<syntaxhighlight lang='bash'> | |||
unzip bootcamp_august_6th_2020.zip | |||
</syntaxhighlight> | |||
* View files | |||
<syntaxhighlight lang='bash'> | |||
cd bootcamp_august_6th_2020 | |||
ls -lrt # long list, in reverse timeline | |||
ls cov_data # a folder of 100 CoV2 genomes in FASTA format, pairwise genome alignment in sam format generated by bwa (or nucmer) | |||
# Indexed sorted bam files generated by samtools | |||
# We skipped bwa (or nucmer) and samtools part of the tutorial for time constrain. | |||
#The bash script "sam-align-genome.bash" used to generate these files is available in the bootcamp_august_6th_2020 folder | |||
ls cov_data/*sorted.bam | wc # 100 sorted.bam files correspond to 100 sequence files | |||
less ref.fas # NC_045512 as reference sequence, "q" to quit | |||
less metadata_cov.txt # a tsv file that contains collection dates and geographic information of 100 CoV2 genomes | |||
* | wc metadata_cov.txt | ||
file TCS.jar # Java application | |||
less bcf-snp-call.sh # a file contain all the bash commands required to call SNPs and generate vcf file of 100 CoV2 genomes | |||
less ploidy.txt # to specify the ploidy=1 during vcf SNP call | |||
less rgb.txt #rgb color code to color the phylogenetic network | |||
</syntaxhighlight> | |||
* Additional files | |||
<syntaxhighlight lang='bash'> | |||
# Output folder: Contains important output files | |||
ls Output | |||
#Use this file in part 2 of the tutorial if you couldn't complete SNP call in part 1 | |||
less Output/cov_haplotypes.nexus | |||
#Use this file in part 3 of the tutorial to make plot if you couldn't complete SNP call in part 1 | |||
less Output/cov_freq_by_continet.txt | |||
</syntaxhighlight> | |||
==Tutorial== | ==Tutorial== | ||
* 2-2:30: Introduction on pathogen phylogenomics | * 2-2:30: Introduction on pathogen phylogenomics | ||
* 2:30-2: | * 2:30-2:55: Part 1: Calling SNPs and creating VCF file | ||
* 2: | |||
* 3: | <syntaxhighlight lang='bash'> | ||
* 3: | ## Create alignment pileup and call variants using plodity file(plodity 1), multiallelic, first output is bam then piped to bcf ## | ||
bcftools mpileup -Ou -f ref.fas cov_data/*sorted.bam | bcftools call -mv --ploidy-file ploidy.txt -Ob -o calls.bcf | |||
## Get Stats, check # of records (SNPs), # of indels, TS/TV ratio | |||
##( expeced more transitions than transversions) #for cov2 should be around more or less 2.5 | |||
bcftools stats calls.bcf | less | |||
#remove indels and save into vcf format | |||
bcftools view --exclude-types indels calls.bcf > snps.vcf | |||
## filter sites by allele counts: only keep informative sites | |||
vcftools --vcf snps.vcf --mac 2 --recode --recode-INFO-all --out snps2.vcf | |||
## rename the snp2 recode file | |||
ls -lrt | |||
mv snps2.vcf.recode.vcf snps2.vcf | |||
#Get stats, check # of records (informative SNPs) and TS/TV ratio (the ratio will increase) | |||
bcftools stats snps2.vcf | less | |||
## Rename the samples | |||
bcftools query -l snps2.vcf > samples | |||
wc samples | |||
less samples | |||
# make sure you don't have exsiting rename_ids.txt file since we are going to append on the file | |||
if [ -e rename_ids.txt ];then rm rename_ids.txt ; fi | |||
cat samples | while read line; do basename $line .sorted.bam >> rename_ids.txt ; done; | |||
wc rename_ids.txt | |||
#change the name of the samples | |||
bcftools reheader -s rename_ids.txt snps2.vcf > snps3.vcf | |||
less snps3.vcf | |||
</syntaxhighlight> | |||
* 2:55-3:10: Part 2: Build and interactive visualize haplotype network with TCS and tcsBU | |||
<syntaxhighlight lang='bash'> | |||
#create fasta file of snps | |||
cat rename_ids.txt | while read line; do echo ">$line"; bcftools query -s "$line" -f '[%TGT]' snps3.vcf; echo; done > cov_haplotypes.fas | |||
grep ">" cov_haplotypes.fas | wc | |||
# Preparing files for haplotype network visualization by parsing the cov metadata | |||
# create file for geographic information | |||
cut -f1,3 metadata_cov.txt | tr '\t' ';' > haplotype.csv | |||
wc haplotype.csv | |||
# Create file to color the network by geography | |||
cut -f3 metadata_cov.txt | sort | uniq > continent.txt | |||
wc continent.txt | |||
paste continent.txt rgb.txt | tr '\t' ';' > groups.csv | |||
less groups.csv | |||
#covert the SNPs fasta file to nexus sequential format | |||
# go to sequence format converter website (find the link in Bioinformatic tools section) | |||
mv result.nexuss cov_haplotypes.nexus | |||
less cov_haplotypes.nexus | |||
#if by any chance you are unable to complete part 1 | |||
#then please use backup "cov_haplotypes.nexus" file inside the "Output" folder for rest of the steps | |||
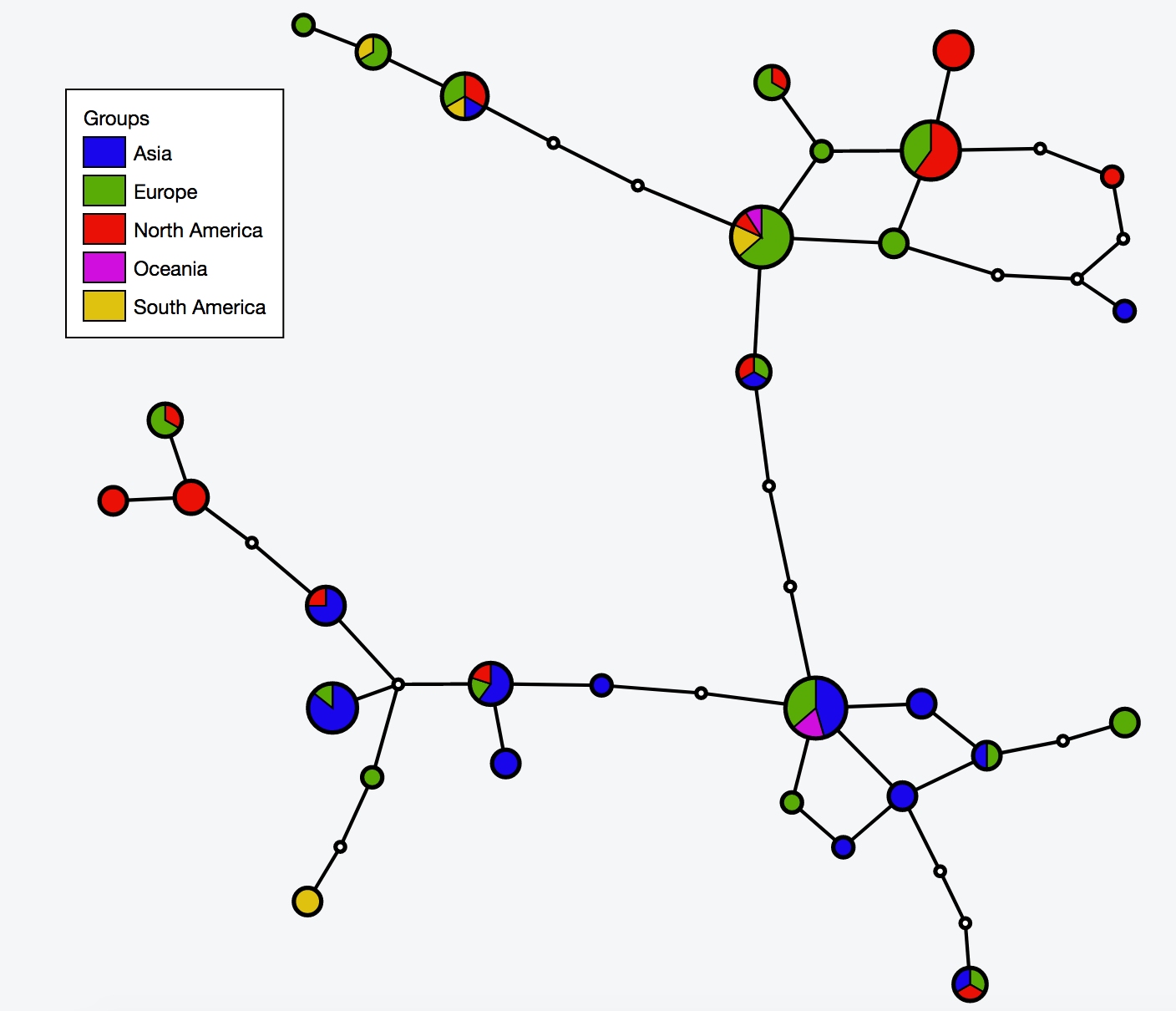
# Build haplotype network for the cov dataset | |||
java -jar -Xmx1g TCS.jar | |||
## This part is not command line. | |||
** A java window will pop-up | |||
** Click on "Start New TCS Analysis" | |||
** Fix connection limit at 5 steps | |||
** File -> Select Nexus/Phylip Sequence file -> upload "cov_haplotypes.nexus" file -> Run | |||
# check the output files | |||
ls -lrt | |||
# Interactive visualization of haplotype network with tcsBU | |||
# go to tcsBU website (find the link in Bioinformatic tools section) | |||
** Load graph file -> cov_haplotypes.nexus.graph | |||
** Load group file -> groups.csv | |||
** Load haplotype file -> haplotype.csv | |||
** Show Legend | |||
** Save SVG | |||
</syntaxhighlight> | |||
{| | |||
|- | |||
| [[File:Cov network.png|thumbnail]] | |||
|} | |||
* 3:10-3:25: Part 3: Visualization of SNPs frequency with vcftools and ggplot2 | |||
<syntaxhighlight lang='bash'> | |||
# calculate the SNPs frequency in Europe and North America | |||
grep -E -- "North America|Europe" metadata_cov.txt | cut -f1 > Europe_NA.ids | |||
wc Europe_NA.ids | |||
bcftools view -S Europe_NA.ids snps3.vcf > Europe_NA.vcf # create Europe and North America subset vcf file | |||
bcftools stats Europe_NA.vcf | less | |||
vcftools --vcf Europe_NA.vcf --freq --out Europe_NA | |||
less Europe_NA.frq | |||
cut -f2,6 Europe_NA.frq | tail -n +2 | sed $'s/.://' > Europe_NA_minor.frq | |||
less Europe_NA_minor.frq | |||
# calculate the SNPs frequency for Asia | |||
grep "Asia" metadata_cov.txt | cut -f1 > Asia.ids | |||
wc Asia.ids | |||
bcftools view -S Asia.ids snps3.vcf > Asia.vcf # create Asia subset vcf file | |||
bcftools stats Asia.vcf | less | |||
vcftools --vcf Asia.vcf --freq --out Asia | |||
less Asia.frq | |||
cut -f2,6 Asia.frq | tail -n +2 | sed $'s/.://' > Asia_minor.frq | |||
less Asia_minor.frq | |||
paste Europe_NA_minor.frq Asia_minor.frq | cut -f1,2,4 > cov_freq_by_continet.txt | |||
less cov_freq_by_continet.txt | |||
wc cov_freq_by_continet.txt | |||
#if by any chance you are unable to complete part 1 | |||
#then please use backup "cov_freq_by_continet.txt" file inside the "Output" folder to make SNPs frequency plot | |||
# SNPs frequency plot by Continent | |||
library(ggplot2) | |||
freq=read.table("cov_freq_by_continet.txt", sep = "\t") | |||
colnames(freq)=c("position", "freq1", "freq2") | |||
head(freq) | |||
ggplot(freq) + | |||
geom_segment( aes(x=position, xend=position, y=freq1, yend=freq2), color="grey") + | |||
geom_point( aes(x=position, y=freq1, color='#EA1006'), size=1.5) + | |||
geom_point( aes(x=position, y=freq2, color='#1A06EA'), size=1.5) + | |||
scale_colour_manual(name = 'Continent', values =c('#EA1006'='#EA1006','#1A06EA'='#1A06EA'), | |||
labels = c('Asia', 'Europe_NA')) + | |||
theme_bw() + xlab("SNPs Position") + ylab("Frequency") + | |||
theme(axis.text.y=element_text(size = 8, face = "bold"), | |||
axis.text.x=element_text(size = 8, face = "bold"), | |||
axis.title=element_text(size=8,face="bold"), | |||
legend.title=element_blank(), legend.text=element_text(size=10 , face = "bold"), | |||
legend.position="top", strip.text.x = element_text(size = 8, face = "bold")) | |||
</syntaxhighlight> | |||
{| | |||
|- | |||
| | |||
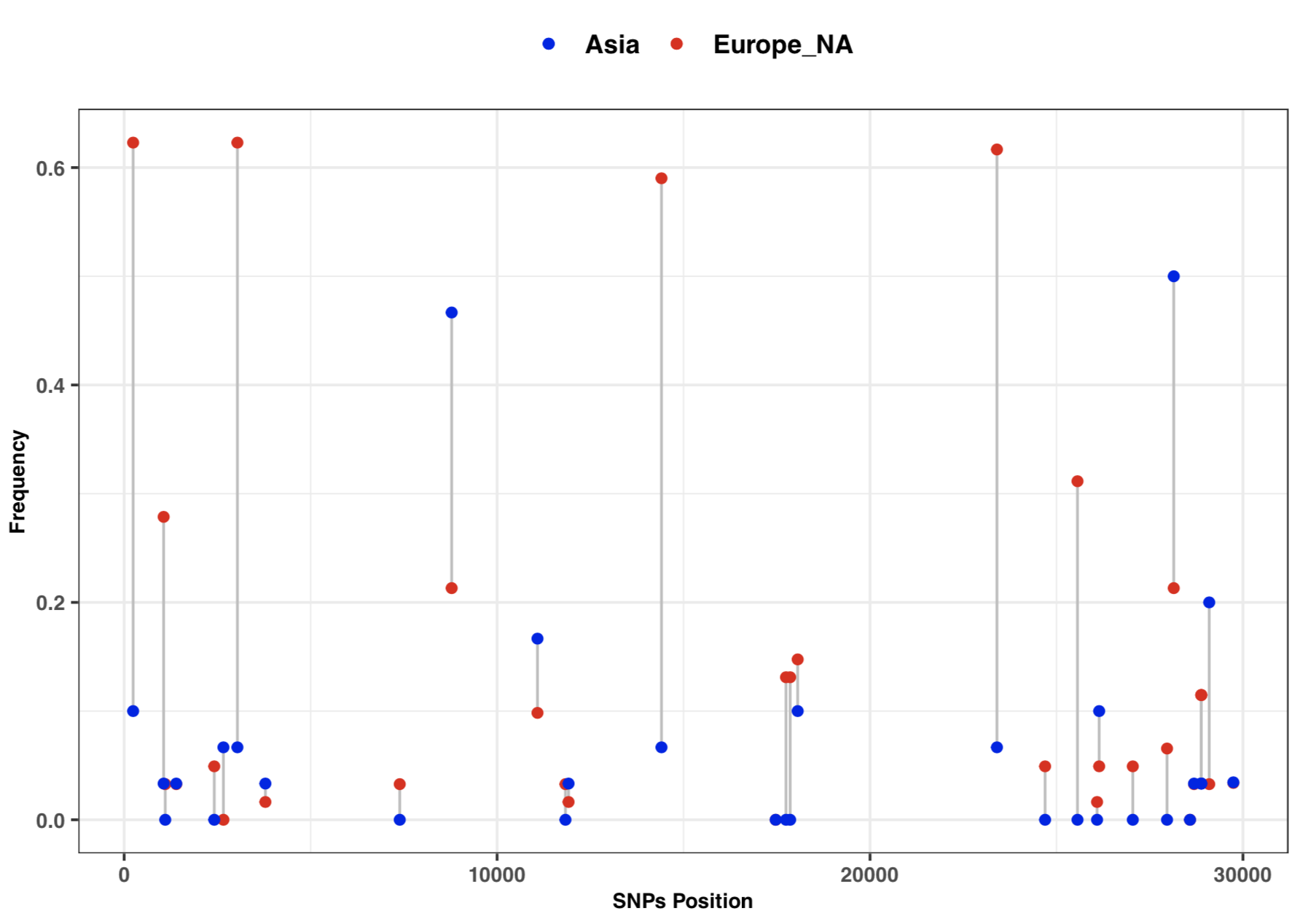
[[File:Cov freq by continent.png |thumbnail]] | |||
|} | |||
* 3:25-3:30: Q & A | |||
Latest revision as of 03:00, 6 August 2020
| CoV Genome Tracker | Coronavirus evolutuon | Lyme Disease (Borreliella) |
|---|---|---|
Case studies
Bioinformatics tools for genomic epidemiology
Required for the tutorial
- bcftools: Reading/writing BCF2/VCF/gVCF files and calling/filtering/summarising SNP and short indel sequence variants Installation link
cd bcftools-1.10.2
./configure
make
make install
## You may need sudo permissions to run make install
bcftools --version
# to check the man page
man bcftools
- vcftools: To work with genetic variation data in the form of VCF files Github link Build from Release Tarball
#download version v0.1.16 from second link
cd vcftools-0.1.16
./configure
make
make install
## You may need sudo permissions to run make install
## Alternative: if you have homebrew in your computer
brew install vcftools
vcftools --version
# to check the man page
man vcftools
- Sequence format converter Web tool
- TCS: To build Haplotype network, TCS.jar file is provided in the dataset folder below, Required Java. PubMed link
- Web-interactive visualization of Haplotype Network with tcsBU Web tool; Paper
- library(ggplot2) (R)
Not required for the tutorial. Recommended
- BpWrapper: command-line tools for manipulation of sequences, alignment, and tree (based on BioPerl). Github Link; Flowchart from publication
- Pairwise genome alignment with MUMMER: Github link
- Samtools: Reading/writing/editing/indexing/viewing SAM/BAM/CRAM format Installation link
CoV genome data set
- N=100 SARS-CoV-2 genomes collected during January, February & March 2020. Data source & acknowledgement GIDAID (Warning: You need to acknowledge GISAID if you reuse the data in any publication)
- Download the folder "bootcamp_august_6th_2020": data file
- unzip the folder
unzip bootcamp_august_6th_2020.zip
- View files
cd bootcamp_august_6th_2020
ls -lrt # long list, in reverse timeline
ls cov_data # a folder of 100 CoV2 genomes in FASTA format, pairwise genome alignment in sam format generated by bwa (or nucmer)
# Indexed sorted bam files generated by samtools
# We skipped bwa (or nucmer) and samtools part of the tutorial for time constrain.
#The bash script "sam-align-genome.bash" used to generate these files is available in the bootcamp_august_6th_2020 folder
ls cov_data/*sorted.bam | wc # 100 sorted.bam files correspond to 100 sequence files
less ref.fas # NC_045512 as reference sequence, "q" to quit
less metadata_cov.txt # a tsv file that contains collection dates and geographic information of 100 CoV2 genomes
wc metadata_cov.txt
file TCS.jar # Java application
less bcf-snp-call.sh # a file contain all the bash commands required to call SNPs and generate vcf file of 100 CoV2 genomes
less ploidy.txt # to specify the ploidy=1 during vcf SNP call
less rgb.txt #rgb color code to color the phylogenetic network
- Additional files
# Output folder: Contains important output files
ls Output
#Use this file in part 2 of the tutorial if you couldn't complete SNP call in part 1
less Output/cov_haplotypes.nexus
#Use this file in part 3 of the tutorial to make plot if you couldn't complete SNP call in part 1
less Output/cov_freq_by_continet.txt
Tutorial
- 2-2:30: Introduction on pathogen phylogenomics
- 2:30-2:55: Part 1: Calling SNPs and creating VCF file
## Create alignment pileup and call variants using plodity file(plodity 1), multiallelic, first output is bam then piped to bcf ##
bcftools mpileup -Ou -f ref.fas cov_data/*sorted.bam | bcftools call -mv --ploidy-file ploidy.txt -Ob -o calls.bcf
## Get Stats, check # of records (SNPs), # of indels, TS/TV ratio
##( expeced more transitions than transversions) #for cov2 should be around more or less 2.5
bcftools stats calls.bcf | less
#remove indels and save into vcf format
bcftools view --exclude-types indels calls.bcf > snps.vcf
## filter sites by allele counts: only keep informative sites
vcftools --vcf snps.vcf --mac 2 --recode --recode-INFO-all --out snps2.vcf
## rename the snp2 recode file
ls -lrt
mv snps2.vcf.recode.vcf snps2.vcf
#Get stats, check # of records (informative SNPs) and TS/TV ratio (the ratio will increase)
bcftools stats snps2.vcf | less
## Rename the samples
bcftools query -l snps2.vcf > samples
wc samples
less samples
# make sure you don't have exsiting rename_ids.txt file since we are going to append on the file
if [ -e rename_ids.txt ];then rm rename_ids.txt ; fi
cat samples | while read line; do basename $line .sorted.bam >> rename_ids.txt ; done;
wc rename_ids.txt
#change the name of the samples
bcftools reheader -s rename_ids.txt snps2.vcf > snps3.vcf
less snps3.vcf
- 2:55-3:10: Part 2: Build and interactive visualize haplotype network with TCS and tcsBU
#create fasta file of snps
cat rename_ids.txt | while read line; do echo ">$line"; bcftools query -s "$line" -f '[%TGT]' snps3.vcf; echo; done > cov_haplotypes.fas
grep ">" cov_haplotypes.fas | wc
# Preparing files for haplotype network visualization by parsing the cov metadata
# create file for geographic information
cut -f1,3 metadata_cov.txt | tr '\t' ';' > haplotype.csv
wc haplotype.csv
# Create file to color the network by geography
cut -f3 metadata_cov.txt | sort | uniq > continent.txt
wc continent.txt
paste continent.txt rgb.txt | tr '\t' ';' > groups.csv
less groups.csv
#covert the SNPs fasta file to nexus sequential format
# go to sequence format converter website (find the link in Bioinformatic tools section)
mv result.nexuss cov_haplotypes.nexus
less cov_haplotypes.nexus
#if by any chance you are unable to complete part 1
#then please use backup "cov_haplotypes.nexus" file inside the "Output" folder for rest of the steps
# Build haplotype network for the cov dataset
java -jar -Xmx1g TCS.jar
## This part is not command line.
** A java window will pop-up
** Click on "Start New TCS Analysis"
** Fix connection limit at 5 steps
** File -> Select Nexus/Phylip Sequence file -> upload "cov_haplotypes.nexus" file -> Run
# check the output files
ls -lrt
# Interactive visualization of haplotype network with tcsBU
# go to tcsBU website (find the link in Bioinformatic tools section)
** Load graph file -> cov_haplotypes.nexus.graph
** Load group file -> groups.csv
** Load haplotype file -> haplotype.csv
** Show Legend
** Save SVG
- 3:10-3:25: Part 3: Visualization of SNPs frequency with vcftools and ggplot2
# calculate the SNPs frequency in Europe and North America
grep -E -- "North America|Europe" metadata_cov.txt | cut -f1 > Europe_NA.ids
wc Europe_NA.ids
bcftools view -S Europe_NA.ids snps3.vcf > Europe_NA.vcf # create Europe and North America subset vcf file
bcftools stats Europe_NA.vcf | less
vcftools --vcf Europe_NA.vcf --freq --out Europe_NA
less Europe_NA.frq
cut -f2,6 Europe_NA.frq | tail -n +2 | sed $'s/.://' > Europe_NA_minor.frq
less Europe_NA_minor.frq
# calculate the SNPs frequency for Asia
grep "Asia" metadata_cov.txt | cut -f1 > Asia.ids
wc Asia.ids
bcftools view -S Asia.ids snps3.vcf > Asia.vcf # create Asia subset vcf file
bcftools stats Asia.vcf | less
vcftools --vcf Asia.vcf --freq --out Asia
less Asia.frq
cut -f2,6 Asia.frq | tail -n +2 | sed $'s/.://' > Asia_minor.frq
less Asia_minor.frq
paste Europe_NA_minor.frq Asia_minor.frq | cut -f1,2,4 > cov_freq_by_continet.txt
less cov_freq_by_continet.txt
wc cov_freq_by_continet.txt
#if by any chance you are unable to complete part 1
#then please use backup "cov_freq_by_continet.txt" file inside the "Output" folder to make SNPs frequency plot
# SNPs frequency plot by Continent
library(ggplot2)
freq=read.table("cov_freq_by_continet.txt", sep = "\t")
colnames(freq)=c("position", "freq1", "freq2")
head(freq)
ggplot(freq) +
geom_segment( aes(x=position, xend=position, y=freq1, yend=freq2), color="grey") +
geom_point( aes(x=position, y=freq1, color='#EA1006'), size=1.5) +
geom_point( aes(x=position, y=freq2, color='#1A06EA'), size=1.5) +
scale_colour_manual(name = 'Continent', values =c('#EA1006'='#EA1006','#1A06EA'='#1A06EA'),
labels = c('Asia', 'Europe_NA')) +
theme_bw() + xlab("SNPs Position") + ylab("Frequency") +
theme(axis.text.y=element_text(size = 8, face = "bold"),
axis.text.x=element_text(size = 8, face = "bold"),
axis.title=element_text(size=8,face="bold"),
legend.title=element_blank(), legend.text=element_text(size=10 , face = "bold"),
legend.position="top", strip.text.x = element_text(size = 8, face = "bold"))
- 3:25-3:30: Q & A