EEB BootCamp 2020: Difference between revisions
Jump to navigation
Jump to search
Bioinformatics Boot Camp for Ecology & Evolution: Genomic Epidemiology
Thursday, Aug 6, 2020, 2 - 3:30pm
Instructors: Dr Weigang Qiu & Ms Saymon Akther
Email: weigang@genectr.hunter.cuny.edu
Lab Website: http://diverge.hunter.cuny.edu/labwiki/
imported>Lab No edit summary |
imported>Lab mNo edit summary |
||
| Line 28: | Line 28: | ||
==Bioinformatics tools for genomic epidemiology== | ==Bioinformatics tools for genomic epidemiology== | ||
Required for the tutorial | Required for the tutorial | ||
* bcftools | * bcftools: Reading/writing BCF2/VCF/gVCF files and calling/filtering/summarising SNP and short indel sequence variants [http://www.htslib.org/download/ Installation link] | ||
* vcftools | * vcftools: To work with genetic variation data in the form of VCF files [https://github.com/vcftools/vcftools Github link] | ||
* TCS: To infer Haplotype network, TCS.jar file is provided, Required Java. [https://pubmed.ncbi.nlm.nih.gov/11050560/ PubMed link] | * TCS: To infer Haplotype network, TCS.jar file is provided, Required Java. [https://pubmed.ncbi.nlm.nih.gov/11050560/ PubMed link] | ||
* Web-interactive visualization of Haplotype Network with tcsBU | * Web-interactive visualization of Haplotype Network with tcsBU [https://cibio.up.pt/software/tcsBU/index.html Web tool]; [https://academic.oup.com/bioinformatics/article/32/4/627/1744448 Paper] | ||
Not required for the tutorial. Recommended | |||
* BpWrapper: command-line tools for manipulation of sequences, alignment, and tree (based on BioPerl). [https://github.com/bioperl/p5-bpwrapper Github Link]; [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-018-2074-9/figures/1 Flowchart from publication] | * BpWrapper: command-line tools for manipulation of sequences, alignment, and tree (based on BioPerl). [https://github.com/bioperl/p5-bpwrapper Github Link]; [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-018-2074-9/figures/1 Flowchart from publication] | ||
* Pairwise genome alignment with MUMMER: [https://github.com/mummer4/mummer Github link] | * Pairwise genome alignment with MUMMER: [https://github.com/mummer4/mummer Github link] | ||
* | * Samtools: Reading/writing/editing/indexing/viewing SAM/BAM/CRAM format [http://www.htslib.org/download/ Installation link] | ||
==CoV genome data set== | ==CoV genome data set== | ||
Revision as of 04:06, 4 August 2020
| CoV Genome Tracker | Coronavirus evolutuon | Lyme Disease (Borreliella) |
|---|---|---|
Case studies
- Is it necessary to mention nextstrain?
Bioinformatics tools for genomic epidemiology
Required for the tutorial
- bcftools: Reading/writing BCF2/VCF/gVCF files and calling/filtering/summarising SNP and short indel sequence variants Installation link
- vcftools: To work with genetic variation data in the form of VCF files Github link
- TCS: To infer Haplotype network, TCS.jar file is provided, Required Java. PubMed link
- Web-interactive visualization of Haplotype Network with tcsBU Web tool; Paper
Not required for the tutorial. Recommended
- BpWrapper: command-line tools for manipulation of sequences, alignment, and tree (based on BioPerl). Github Link; Flowchart from publication
- Pairwise genome alignment with MUMMER: Github link
- Samtools: Reading/writing/editing/indexing/viewing SAM/BAM/CRAM format Installation link
CoV genome data set
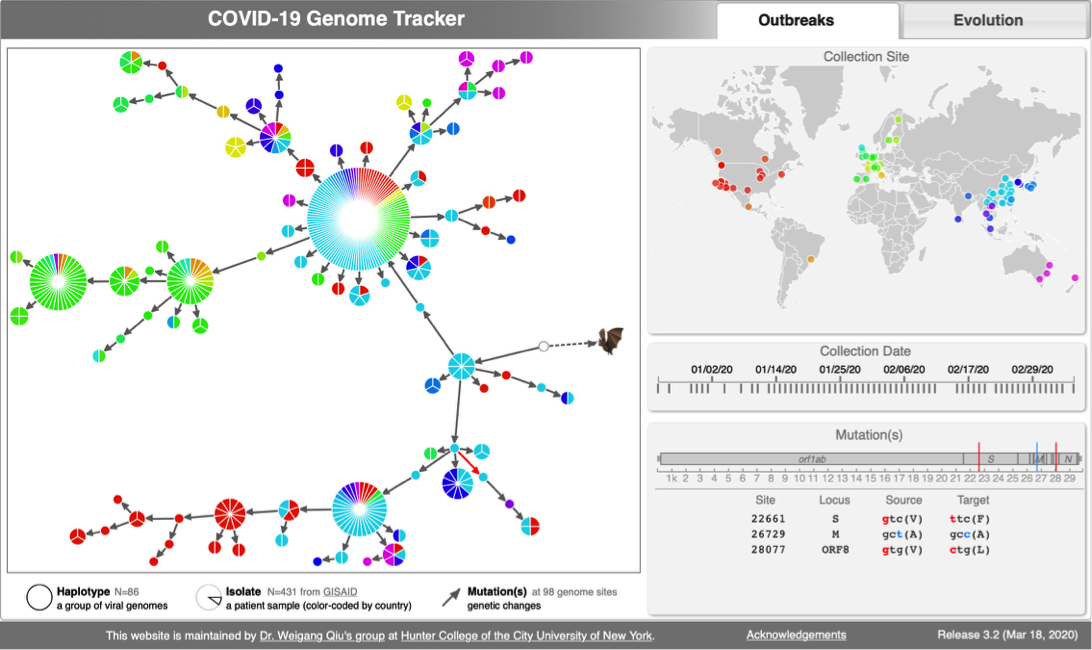
- N=100 SARS-CoV-2 genomes collected during January, February & March 2020. Data source & acknowledgement GIDAID (Warning: You need to acknowledge GISAID if you reuse the data in any publication)
- Download the folder "bootcamp_august_6th_2020": data file
- unzip the folder
unzip bootcamp_august_6th_2020.zip
- View files
ls -lrt # long list, in reverse timeline
ls cov_data # a folder of 100 CoV2 genomes in FASTA format, pairwise genome alignment sam and indexed sorted bam files generated by bwa (or nucmer) and samtools
# We skipped bwa (or nucmer) and samtools part of the tutorial for time constrain. The bash script used to generate these files is available on request
ls cov_data/*sorted.bam | wc # 100 sorted.bam files correspond to 100 sequence files
less ref.fas # NC_045512 as reference sequence, "q" to quit
less metadata_cov.txt # a tsv file that contains collection dates and geographic information of 100 CoV2 genomes
wc metadata_cov.txt
file TCS.jar # Java application
less bcf-snp-call.sh # a file contain all the bash commands required to call SNPs and generate vcf file of 100 CoV2 genomes
less ploidy.txt # to specify the ploidy=1 during vcf SNP call
less rgb.txt #rgb color code to color the phylogenetic network
Tutorial
- 2-2:30: Introduction on pathogen phylogenomics
- 2:30-2:45: Demo: sequence manipulation with BpWrapper
bioseq --man
bioseq -i'genbank' ref.gb > ref.fas
bioseq -n Jan-Feb.mafft
bioaln --man
bioaln -n -i'fasta' Jan-Feb.mafft
bioaln -l -i'fasta' Jan-Feb.mafft
bioaln -n -i'phylip' cov-565strains-617snvs.phy
bioaln -l -i'phylip' cov-565strains-617snvs.phy
FastTree -nt cov-565strains-617snvs.phy > cov.dnd
biotree --man
biotree -n cov.dnd
biotree -l cov.dnd
- 2:45-3:10: build haplotype network with TCS
# Data pre-processing
# 1. Download genomes & meta data from GISAID
# 2. Run dnadist against a reference genome
man nucmer
dnadiff -h
dnadiff ref.fas <query FASTA>
mkdir fasta-files
cd fasta-files
for f in *.fas; do dnadiff ref.fas $f; done
<to be added: plot in R seq diff vs collection date>
# 3. Remove mis-assembled and reverse-complemented genomes
bioseq -d'file:'
# 4. Remove genomes with more than 10 non-ATCG bases
bioseq -d'ambig:10'
# 5. Run mafft (not run; takes too long)
# 6. Run snp-sites
snp-sites
java -jar -Xmx1g TCS.jar
- 3:10-3:20: interactive visualization with BuTCS
- Load graph file
- Load group file
- Load haplotype file
- 3:20-3:30: Q & A