Bioutils: Difference between revisions
imported>Weigang |
imported>Weigang |
||
| (32 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<span style="color: Seagreen;font-weight:bold;font-size:large;"><u> | <span style="color: Seagreen;font-weight:bold;font-size:large;"><u>B</u>io<u>P</u>erl-based Sequence <u>Util</u>ities<span> | ||
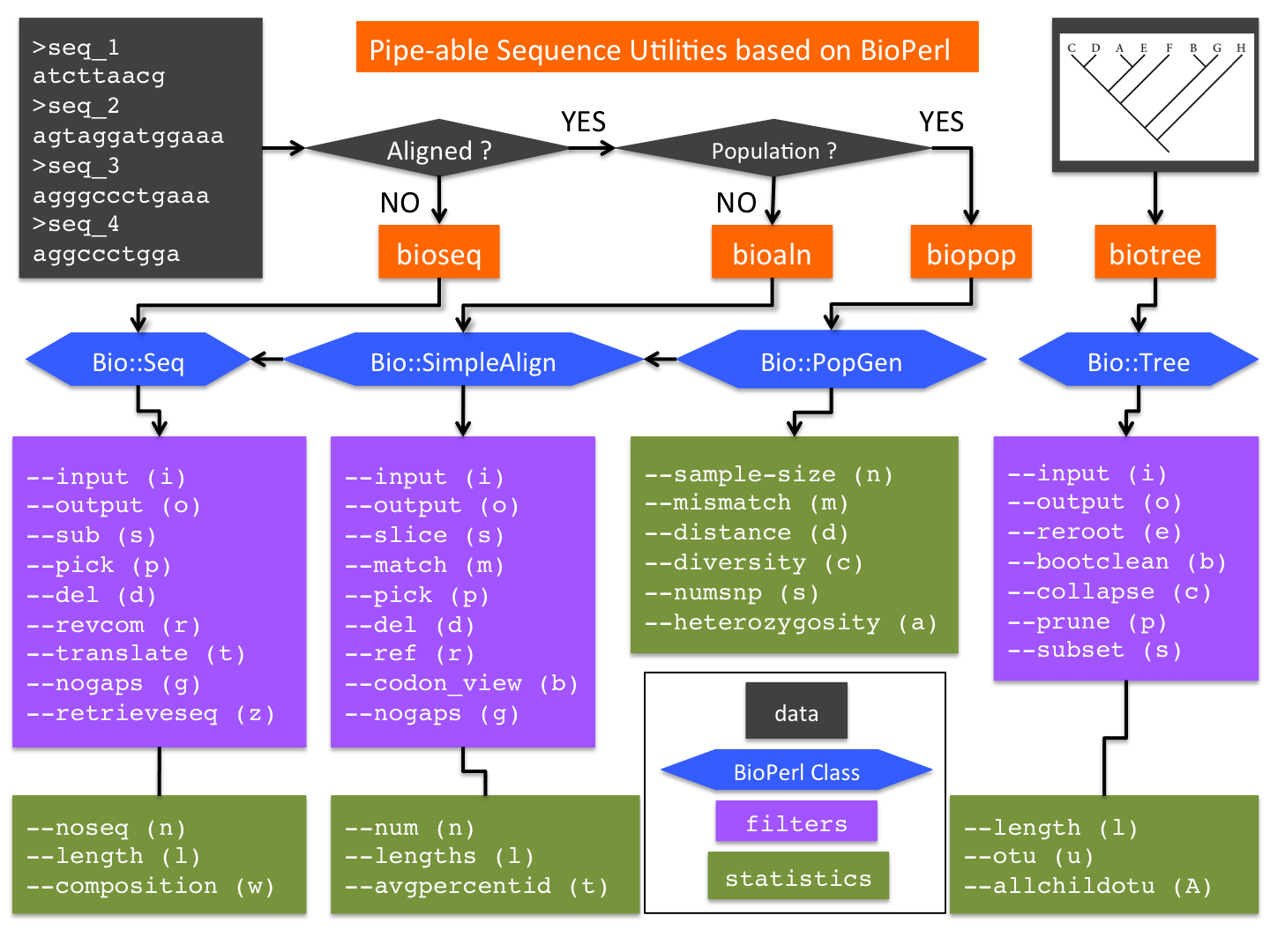
[[File:Seq-util.png|thumbnail|600px|right|'''Figure 1'''. Design and Methods of '' | [[File:Seq-util.png|thumbnail|600px|right|'''Figure 1'''. Design and Methods of ''bputils'']] | ||
==What is '' | ==What is ''bputils''?== | ||
'' | ''bputils'' is a suite of Perl scripts that provide convenient command-line access to popular BioPerl methods. Designed as UNIX utilities, these tools aim to circumvent a constant need (and urge) to compose one-off BioPerl scripts for routine manipulations of sequences, alignments and trees. | ||
The initial release of '' | The initial release of ''bputils'' consists of four inter-connected utilities ('''Figure 1'''): | ||
* '''bioseq''': a wrapper of BioPerl class Bio::Seq (with additional methods) | * '''bioseq''': a wrapper of BioPerl class Bio::Seq (with additional methods) | ||
* '''bioaln''': a wrapper of Bio::SimpleAlign (which inherits Bio::Seq; with additional methods) | * '''bioaln''': a wrapper of Bio::SimpleAlign (which inherits Bio::Seq; with additional methods) | ||
| Line 10: | Line 10: | ||
* '''biotree''': a wrapper of Bio::tree (with additional methods) | * '''biotree''': a wrapper of Bio::tree (with additional methods) | ||
These utilities have been in development since 2002 in [[Main Page|the lab of Dr Weigang Qiu]] at Hunter College of the City University of New York. They are the main code base of the Qiu Lab, which specializes in microbial evolutionary genomics. They proved to be convenient, efficient, and popular among students and researchers passing through the lab. By releasing ''bioutils'' as an Open Source tool (perhaps as a part of bioperl distribution), we hope to (1) share our | These utilities have been in development since 2002 in [[Main Page|the lab of Dr Weigang Qiu]] at Hunter College of the City University of New York. They are the main code base of the Qiu Lab, which specializes in microbial evolutionary genomics. They proved to be convenient, efficient, and popular among students and researchers passing through the lab. By releasing ''bioutils'' as an Open Source tool (perhaps as a part of bioperl distribution), we hope to (1) share our tools and (2) invite other developers to join the effort of making BioPerl more accessible. | ||
---- | ---- | ||
==Other Unix utilities for genomics== | |||
* [http://www.bioperl.org/wiki/Bioperl_scripts BioPerl scripts] (This is probably where these utilities will eventually be housed) | |||
* EMBOSS: [http://emboss.sourceforge.net/apps/ Official site]; [http://manuals.bioinformatics.ucr.edu/home/emboss a third-party manual] | |||
* NEWICK Utilities: [http://cegg.unige.ch/newick_utils Official website]; [http://www.ncbi.nlm.nih.gov/pubmed/20472542 A publication] | |||
==Live Demos== | ==Live Demos== | ||
| Line 23: | Line 27: | ||
bioseq -t6 foo.fasta # translate in all 6 frames | bioseq -t6 foo.fasta # translate in all 6 frames | ||
bioseq -p'id:seq_1' foo.fasta # pick a sequence by ID | bioseq -p'id:seq_1' foo.fasta # pick a sequence by ID | ||
bioseq -p'order:3' # pick the 3rd sequence | bioseq -p'order:3' foo.fasta # pick the 3rd sequence | ||
bioseq -p're:Human' foo.fasta # pick all sequences labeled as "Human" (by regular expression) | bioseq -p're:Human' foo.fasta # pick all sequences labeled as "Human" (by regular expression) | ||
bioseq -g foo.fasta # remove | bioseq -g foo.fasta # remove gaps | ||
bioseq -z'CP003201' -o'genbank' # retrieve a GenBank file with accession | bioseq -z'CP003201' -o'genbank' # retrieve a GenBank file with accession | ||
bioseq -z'CP003201' -o'fasta' # same file in FASTA | bioseq -z'CP003201' -o'fasta' # same file in FASTA | ||
| Line 31: | Line 35: | ||
* bioaln | * bioaln | ||
<syntaxhighlight lang=bash"> | <syntaxhighlight lang=bash"> | ||
bioaln -i'fasta' -o'phylip' foo.fasta # convert | bioaln -i'fasta' -o'phylip' foo.fasta # convert FASTA alignment to PHYLIP | ||
bioaln -l foo.aln # print alignment length of a CLUSTALW (default format) file | bioaln -l foo.aln # print alignment length of a CLUSTALW (default format) file | ||
bioaln -s'100, 200' foo.aln # obtain an alignment slice | bioaln -s'100, 200' foo.aln # obtain an alignment slice | ||
bioaln -m foo.aln # show only variable sites | bioaln -m foo.aln # show only variable sites | ||
bioaln -r'seq_2' foo.aln # | bioaln -r'seq_2' foo.aln # set "seq_2" as reference (first) sequence | ||
bioaln -g foo.aln # remove gapped sites | bioaln -g foo.aln # remove gapped sites | ||
bioaln -p'seq_1,seq_3,seq_6' foo.aln # pick a subset of sequences | bioaln -p'seq_1,seq_3,seq_6' foo.aln # pick a subset of sequences | ||
| Line 42: | Line 46: | ||
* biotree | * biotree | ||
<syntaxhighlight lang=bash"> | <syntaxhighlight lang=bash"> | ||
biotree -l foo.newick # total tree length | |||
biotree -e 'outgroup' foo.newick # re-root on a "outgroup" | |||
biotree -z foo.newick # print pair-wise tree distances between leafs | |||
biotree -p 'node1' -p 'node2' foo.newick # remove "node1" and node2" | |||
biotree -s 'node1' -s 'node2' -s 'node3' -s'node4' foo.newick # subtree consisting ONLY of these nodes | |||
</syntaxhighlight> | </syntaxhighlight> | ||
* biopop | * biopop | ||
<syntaxhighlight lang=bash"> | <syntaxhighlight lang=bash"> | ||
biopop --stats 'pi,theta' foo.aln # print pi and theta of population (default: CLUSTALW alignment) | |||
biopop -m foo.aln # print mismatch distribution | |||
biopop -d foo.aln # print distance matrix (default: Jukes-Cantor model; should be back-tracked into "bioaln") | |||
biopop -s foo.aln # obtain number of segregating sites (i.e., SNPs) | |||
biopop -a foo.aln # get heterozygosity for each SNP site | |||
</syntaxhighlight> | </syntaxhighlight> | ||
===Power usage (with pipes)=== | ===Power usage (with pipes)=== | ||
<syntaxhighlight lang=bash"> | <syntaxhighlight lang=bash"> | ||
bioseq -p'order:5' foo.fasta | bioseq -s'100,200' | bioseq -r | bioseq -t1 # pick, subseq, revcom, and translate | bioseq -p'order:5' foo.fasta | bioseq -s'100,200' | bioseq -r | bioseq -t1 # pick, subseq, revcom, and translate | ||
bioaln -o'fasta' foo.aln | bioseq -g # remove gaps within individual sequences | bioaln -o'fasta' foo.aln | bioseq -g # remove gaps within individual sequences | ||
bioaln -o'fasta' foo.aln | bioseq -t1 | bioaln -i'fasta' # turn a nucleotide alignment into a peptide alignment | |||
biotree -s'otu1' -s'otu2' -s'otu3' foo.newick | biotree -l # subset a large tree and get total tree length | |||
</syntaxhighlight> | </syntaxhighlight> | ||
===Creative usage (with BASH | |||
===Creative usage (with BASH)=== | |||
<syntaxhighlight lang=bash"> | <syntaxhighlight lang=bash"> | ||
echo -ne ">lookup\nATG\n" | bioseq -t1 # Lookup a codon product | echo -ne ">lookup\nATG\n" | bioseq -t1 # Lookup a codon product | ||
len=$(bioaln -l foo.aln); len_degap=$(bioaln -g foo.aln | bioaln -l); echo "$len-$len_degap" | bc -l # count alignment gaps | len=$(bioaln -l foo.aln); len_degap=$(bioaln -g foo.aln | bioaln -l); echo "$len-$len_degap" | bc -l # count alignment gaps | ||
for i in {1..100}; do bioaln --boot foo.aln >> foo.boot; done # bootstrap an alignment (with replacement) | |||
</syntaxhighlight> | |||
===Even-more powerful usage (with other applications)=== | |||
<syntaxhighlight lang=bash"> | |||
# Permutation Trait Probability test for tree-ness: | |||
for i in {1..100); do | |||
bioaln -P -i'fasta' -o'fasta' nt.fas > nt.permuted.fas; | |||
FastTree -nt nt.permuted.fas 2> /dev/null | biotree -l >> tree-length.txt # tree length of permuted alignment | |||
done; | |||
</syntaxhighlight> | </syntaxhighlight> | ||
---- | ---- | ||
==Full documentation== | ==Full documentation== | ||
<div class=" | <div class="mw-collapsible mw-collapsed" style="width:100%"> | ||
POD for bioseq | POD document for '''bioseq''' | ||
<div class="mw-collapsible-content | <div class="mw-collapsible-content"> | ||
* NAME | |||
<p>bioseq - Fasta sequence editing module based on BioPerl.</p> | <p>bioseq - Fasta sequence editing module based on BioPerl.</p> | ||
* SYNOPSIS | |||
<p><b>bioseq</b> [options] [sequence file]</p> | <p><b>bioseq</b> [options] [sequence file]</p> | ||
* DESCRIPTION | |||
<p><b>bioseq</b> will read a sequence file and act upon it by doing the following - reformat input (default is fasta) to Genbank or EMBL formats, delete specified sequences, generate overlapping subsequence with a specified window size, generate the reverese complementary sequence, for nucleic acid sequences only, take input list of sequences apart into individual sequence files, extract a specified subset of the sequence, linearize the sequence, remove gaps, find the longest open reading frame (ORF), remove stop codons, give percentage composition of specified amino acids or nuclic acid bases, split the sequences as specified by the user, translate a specific frame of input sequence, or extract a specific gene ID from multiple file sequences. By default, <b>bioseq</b> will assume that both the input and the output are in FASTA format.</p> | <p><b>bioseq</b> will read a sequence file and act upon it by doing the following - reformat input (default is fasta) to Genbank or EMBL formats, delete specified sequences, generate overlapping subsequence with a specified window size, generate the reverese complementary sequence, for nucleic acid sequences only, take input list of sequences apart into individual sequence files, extract a specified subset of the sequence, linearize the sequence, remove gaps, find the longest open reading frame (ORF), remove stop codons, give percentage composition of specified amino acids or nuclic acid bases, split the sequences as specified by the user, translate a specific frame of input sequence, or extract a specific gene ID from multiple file sequences. By default, <b>bioseq</b> will assume that both the input and the output are in FASTA format.</p> | ||
* OPTIONS | |||
<dl> | <dl> | ||
<dt><b>--help, -h</b></dt> | |||
<dt | |||
<dd> | <dd> | ||
<p>Print a brief help message and exit.</p> | <p>Print a brief help message and exit.</p> | ||
Usage: bioseq -h <keyword> | |||
</dd> | </dd> | ||
<dt | <dt><b>--man, -m</b></dt> | ||
<dd> | <dd> | ||
<p>Prints the manual page and exit.</p> | <p>Prints the manual page and exit.</p> | ||
Usage: bioseq -m <keyword> | |||
</dd> | </dd> | ||
<dt | <dt><b>--input, -i</b> 'format'</dt> | ||
<dd> | <dd> | ||
<p>Input file format. By default, this is 'fasta'. For Genbank format, use 'genbank'. For EMBL format, use 'embl'.</p> | <p>Input file format. By default, this is 'fasta'. For Genbank format, use 'genbank'. For EMBL format, use 'embl'.</p> | ||
Usage: bioseq -i 'genbank' input_file | |||
</dd> | </dd> | ||
<dt id="output--o-format"><b>--output, -o</b> 'format'</dt> | <dt id="output--o-format"><b>--output, -o</b> 'format'</dt> | ||
<dd> | <dd> | ||
<p>Output file format. By default, this is 'fasta'.For Genbank format, use 'genbank'. For EMBL format, use 'embl'.</p> | <p>Output file format. By default, this is 'fasta'.For Genbank format, use 'genbank'. For EMBL format, use 'embl'.</p> | ||
Usage: bioseq -o 'EMBL' input_file | |||
</dd> | </dd> | ||
<dt id="noseq--n"><b>--noseq, -n</b></dt> | <dt id="noseq--n"><b>--noseq, -n</b></dt> | ||
<dd> | <dd> | ||
<p>Print number of sequences specified by n.</p> | <p>Print number of sequences specified by n.</p> | ||
Usage: bioseq -n input_file | |||
</dd> | </dd> | ||
<dt id="sub--s-min-max"><b>--sub, -s</b> 'min,max'</dt> | <dt id="sub--s-min-max"><b>--sub, -s</b> 'min,max'</dt> | ||
<dd> | <dd> | ||
<p>Select substring (of 1st sequence),</p> | <p>Select substring (of 1st sequence),</p> | ||
Usage: bioseq -s '<beginning index>, <ending index>' input_file | |||
Example: bioseq -s'20,80' input_file (or --sub'20,80' or -s='20,80' or --sub='20,80') | |||
Example: bioseq -s'20,80' input_file (or --sub'20,80' or -s='20,80' or --sub='20,80') | |||
</dd> | </dd> | ||
<dt id="lengths--l"><b>--lengths, -l</b></dt> | <dt id="lengths--l"><b>--lengths, -l</b></dt> | ||
<dd> | <dd> | ||
<p>Print all sequence lengths.</p> | <p>Print all sequence lengths.</p> | ||
Usage: bioseq -l | |||
</dd> | </dd> | ||
<dt id="leadgaps--y"><b>--leadgaps, -y</b></dt> | <dt id="leadgaps--y"><b>--leadgaps, -y</b></dt> | ||
<dd> | <dd> | ||
<p>Count and return the number of leading gaps in each sequence.</p> | <p>Count and return the number of leading gaps in each sequence.</p> | ||
Usage: bioseq -y | |||
</dd> | </dd> | ||
<dt id="pick--p-tag:value"><b>--pick, -p</b> 'tag:value'</dt> | <dt id="pick--p-tag:value"><b>--pick, -p</b> 'tag:value'</dt> | ||
<dd> | <dd> | ||
<p>Select a single sequence or a comma-separated list of sequences, e.g, --pick 'id:foo' by id --pick 'order:2' by order --pick 're:REGEX' using a regular expression (only one regex is expected)</p> | <p>Select a single sequence or a comma-separated list of sequences, e.g, --pick 'id:foo' by id --pick 'order:2' by order --pick 're:REGEX' using a regular expression (only one regex is expected)</p> | ||
<p>Specifically for a list of sequences, --pick 'id:foo,bar' list by id --pick 'order:2,3' list by order --pick 'order:2-10' list by range</p> | <p>Specifically for a list of sequences, --pick 'id:foo,bar' list by id --pick 'order:2,3' list by order --pick 'order:2-10' list by range</p> | ||
Usage: bioseq -p 'id:<number>' input_file | |||
</dd> | </dd> | ||
<dt id="del--d-tag:value"><b>--del, -d</b> 'tag:value'</dt> | <dt id="del--d-tag:value"><b>--del, -d</b> 'tag:value'</dt> | ||
<dd> | <dd> | ||
Delete a sequence or a comma-separated list of sequences, eg, | |||
--del 'id:foo' by id | --del 'id:foo' by id | ||
--del 'order:2' by order | --del 'order:2' by order | ||
| Line 187: | Line 157: | ||
--del 're:REGEX' using a regular expression (only one regex is expected) | --del 're:REGEX' using a regular expression (only one regex is expected) | ||
Usage: bioseq --del 'id:<number>' input_file | Usage: bioseq --del 'id:<number>' input_file | ||
</dd> | </dd> | ||
| Line 195: | Line 165: | ||
<p>Reverse complement</p> | <p>Reverse complement</p> | ||
Usage: bioseq -r input_file | |||
</dd> | </dd> | ||
| Line 203: | Line 173: | ||
<p>Generate overlapping subsequence with a specified window size (default 1)</p> | <p>Generate overlapping subsequence with a specified window size (default 1)</p> | ||
Usage: bioseq -k '<index of subsequence>' input_file | |||
</dd> | </dd> | ||
| Line 211: | Line 181: | ||
<p>Translate in 1, 3, or 6 frames. eg, -t1, -t3, or -t6.</p> | <p>Translate in 1, 3, or 6 frames. eg, -t1, -t3, or -t6.</p> | ||
Usage: bioseq -t3 input_file | |||
</dd> | </dd> | ||
| Line 219: | Line 189: | ||
<p>Shred into individual sequences</p> | <p>Shred into individual sequences</p> | ||
Usage: bioseq -c input_file | |||
</dd> | </dd> | ||
| Line 227: | Line 197: | ||
<p>Linearize FASTA.</p> | <p>Linearize FASTA.</p> | ||
Usage: bioseq -L fasta_file | |||
</dd> | </dd> | ||
| Line 235: | Line 205: | ||
<p>Extract in-frame sequences.</p> | <p>Extract in-frame sequences.</p> | ||
Usage: bioseq -e input_file | |||
</dd> | </dd> | ||
| Line 243: | Line 213: | ||
<p>Remove gaps</p> | <p>Remove gaps</p> | ||
Usage: bioseq -g input_file | |||
</dd> | </dd> | ||
| Line 251: | Line 221: | ||
<p>Output the frame that gives the longest ORF.</p> | <p>Output the frame that gives the longest ORF.</p> | ||
Usage: bioseq -C input_file | |||
</dd> | </dd> | ||
| Line 259: | Line 229: | ||
<p>Remove stop codons (for e.g., PAML input)</p> | <p>Remove stop codons (for e.g., PAML input)</p> | ||
Usage: bioseq -x input_file | |||
</dd> | </dd> | ||
| Line 267: | Line 237: | ||
<p>Replace sequence IDs with serial IDs 'n' characters long, including a leading 'S' (e.g., -a'5' gives S0001). Produces a sed script file with a '.sed' suffix that may be used with sed's '-f' argument. If the filename is '-', the sed file is named STDOUT.sed instead. The sed filename is specified on STDERR.</p> | <p>Replace sequence IDs with serial IDs 'n' characters long, including a leading 'S' (e.g., -a'5' gives S0001). Produces a sed script file with a '.sed' suffix that may be used with sed's '-f' argument. If the filename is '-', the sed file is named STDOUT.sed instead. The sed filename is specified on STDERR.</p> | ||
Usage: bioseq -a <number> input_file | |||
</dd> | </dd> | ||
| Line 277: | Line 247: | ||
<p>If there are enough sequences that the length of the prefix plus the length of the digit portion exceeds the length given to --anonymize, a warning will be given: aln-manipulations.pl -a 4 --prefix=SEQ # output SEQ1 atg... ... SEQ10 atc...</p> | <p>If there are enough sequences that the length of the prefix plus the length of the digit portion exceeds the length given to --anonymize, a warning will be given: aln-manipulations.pl -a 4 --prefix=SEQ # output SEQ1 atg... ... SEQ10 atc...</p> | ||
# more output | |||
WARNING: Anonymized ID length exceeded requested length: try a different length or prefix. | WARNING: Anonymized ID length exceeded requested length: try a different length or prefix. | ||
</dd> | </dd> | ||
| Line 286: | Line 256: | ||
<p>Base or AA composition.</p> | <p>Base or AA composition.</p> | ||
Usage: bioseq -w input_file | |||
</dd> | </dd> | ||
| Line 294: | Line 264: | ||
<p>Split the input sequence file into several smaller files with 'split_at' sequences per file. For instance, to split a file into several smaller files with 100000 sequences each, you would run:</p> | <p>Split the input sequence file into several smaller files with 'split_at' sequences per file. For instance, to split a file into several smaller files with 100000 sequences each, you would run:</p> | ||
bioseq -S 100000 seq.fasta | |||
<p>The output files will be named with the label "split_N" according to the input file (or with the STDIN prefix if the file is read via standard in), where N denotes the "part" or "split" number.</p> | <p>The output files will be named with the label "split_N" according to the input file (or with the STDIN prefix if the file is read via standard in), where N denotes the "part" or "split" number.</p> | ||
| Line 302: | Line 272: | ||
<dd> | <dd> | ||
Usage: bioseq -z [Accession] | |||
Retrieves the sequence from GenBank using the provided GenBank accession. Prints out text in a fasta file. | Retrieves the sequence from GenBank using the provided GenBank accession. Prints out text in a fasta file. | ||
| Line 322: | Line 292: | ||
AAAACACATGGGACTAACGATAAGGGTGCTAAAGAACTTAAAGAGTTATCTGAATCAGTA | AAAACACATGGGACTAACGATAAGGGTGCTAAAGAACTTAAAGAGTTATCTGAATCAGTA | ||
GAAAGCTTGGCAAAAGCAGCTCAAGCAGCATTAGCTAATTCAGTTAAAGAGCTTACAAGT | GAAAGCTTGGCAAAAGCAGCTCAAGCAGCATTAGCTAATTCAGTTAAAGAGCTTACAAGT | ||
CCTGTTGTGGCAGAAAGTCCAAAAAAACCTTAA | CCTGTTGTGGCAGAAAGTCCAAAAAAACCTTAA | ||
</dd> | </dd> | ||
| Line 330: | Line 300: | ||
<p>Extract two sequences from input file and generate a dotplot.</p> | <p>Extract two sequences from input file and generate a dotplot.</p> | ||
Usage: bioseq -D 'id1,id2,window_size,slider' fasta_file | |||
<p>Id1 and Id2 are extracted with their corresponding sequences. Be sure to use the entire sequence identifer, as this is a whole string match. Window_size corresponds to the number of character you would like to compare (Default window is 10). Slider is the number of windows to compare (Default slider is 10). The sequence corresponding to ID1 will appear on the X axis (row) and ID2 on the Y axis (column). This method will work on both DNA and Amino Acids.</p> | <p>Id1 and Id2 are extracted with their corresponding sequences. Be sure to use the entire sequence identifer, as this is a whole string match. Window_size corresponds to the number of character you would like to compare (Default window is 10). Slider is the number of windows to compare (Default slider is 10). The sequence corresponding to ID1 will appear on the X axis (row) and ID2 on the Y axis (column). This method will work on both DNA and Amino Acids.</p> | ||
Example: | |||
Sample Input: | Sample Input: | ||
>id1 | >id1 | ||
| Line 357: | Line 327: | ||
A | A | ||
T | T | ||
G | G | ||
<p> | <p><b>--rename, -R</b> 'rename_id'</p> | ||
Usage: bioseq -R [file_with_new_names] file_to_be_changed.fasta | |||
Ex: bioseq -D list.txt test-bioseq.nuc | Ex: bioseq -D list.txt test-bioseq.nuc | ||
| Line 389: | Line 359: | ||
>B31 | >B31 | ||
AATTTTTAAAATATAATATAAAAACAGCTAATCCAATAGAAAAATTTTAAAACTTTTCTA | AATTTTTAAAATATAATATAAAAACAGCTAATCCAATAGAAAAATTTTAAAACTTTTCTA | ||
TTGGATAGATTTTATACAAAGAAGGTAATA | TTGGATAGATTTTATACAAAGAAGGTAATA | ||
</dd> | </dd> | ||
</dl> | </dl> | ||
* EXAMPLES | |||
<p>Section under construction...</p> | <p>Section under construction...</p> | ||
* REQUIRES | |||
<p>Perl 5.010, BioPerl</p> | <p>Perl 5.010, BioPerl</p> | ||
* SEE ALSO | |||
perl(1) | |||
* AUTHORS | |||
** Weigang Qiu at genectr.hunter.cuny.edu | |||
** Yözen Hernández yzhernand at gmail dot com | |||
** Levy Vargas levy dot vargas at gmail dot com | |||
</div> | </div> | ||
</div> | </div> | ||
<div class=" | <div class="mw-collapsible mw-collapsed" style="width:100%"> | ||
POD for bioaln | POD document for '''bioaln''' | ||
<div class="mw-collapsible-content"> | <div class="mw-collapsible-content">This text should be hidden by default.</div> | ||
</div> | </div> | ||
<div class=" | <div class="mw-collapsible mw-collapsed" style="width:100%"> | ||
POD for biopop | POD document for '''biopop''' | ||
<div class="mw-collapsible-content"> | <div class="mw-collapsible-content">This text should be hidden by default.</div> | ||
</div> | </div> | ||
<div class=" | <div class="mw-collapsible mw-collapsed" style="width:100%"> | ||
POD for | POD document for '''biopop''' | ||
<div class="mw-collapsible-content"> | <div class="mw-collapsible-content">This text should be hidden by default.</div> | ||
</div> | </div> | ||
---- | ---- | ||
| Line 452: | Line 405: | ||
---- | ---- | ||
== | ==Contributors== | ||
* Yozen Hernandez | * Yozen Hernandez | ||
* Levy Vargas | * Levy Vargas | ||
Latest revision as of 20:47, 8 October 2014
BioPerl-based Sequence Utilities
What is bputils?
bputils is a suite of Perl scripts that provide convenient command-line access to popular BioPerl methods. Designed as UNIX utilities, these tools aim to circumvent a constant need (and urge) to compose one-off BioPerl scripts for routine manipulations of sequences, alignments and trees.
The initial release of bputils consists of four inter-connected utilities (Figure 1):
- bioseq: a wrapper of BioPerl class Bio::Seq (with additional methods)
- bioaln: a wrapper of Bio::SimpleAlign (which inherits Bio::Seq; with additional methods)
- biopop: a wrapper of Bio::PopGen (which can be converted from Bio::SimpleAlign; with additional methods)
- biotree: a wrapper of Bio::tree (with additional methods)
These utilities have been in development since 2002 in the lab of Dr Weigang Qiu at Hunter College of the City University of New York. They are the main code base of the Qiu Lab, which specializes in microbial evolutionary genomics. They proved to be convenient, efficient, and popular among students and researchers passing through the lab. By releasing bioutils as an Open Source tool (perhaps as a part of bioperl distribution), we hope to (1) share our tools and (2) invite other developers to join the effort of making BioPerl more accessible.
Other Unix utilities for genomics
- BioPerl scripts (This is probably where these utilities will eventually be housed)
- EMBOSS: Official site; a third-party manual
- NEWICK Utilities: Official website; A publication
Live Demos
Basic Usage
- bioseq
bioseq -l foo.fasta # print seq names and lengths from FASTA (default format) file
bioseq -r foo.fasta # reverse complement
bioseq -t1 foo.fasta # translate in the +1 frame
bioseq -t3 foo.fasta # translate in +1, +2, and +3 frames
bioseq -t6 foo.fasta # translate in all 6 frames
bioseq -p'id:seq_1' foo.fasta # pick a sequence by ID
bioseq -p'order:3' foo.fasta # pick the 3rd sequence
bioseq -p're:Human' foo.fasta # pick all sequences labeled as "Human" (by regular expression)
bioseq -g foo.fasta # remove gaps
bioseq -z'CP003201' -o'genbank' # retrieve a GenBank file with accession
bioseq -z'CP003201' -o'fasta' # same file in FASTA- bioaln
bioaln -i'fasta' -o'phylip' foo.fasta # convert FASTA alignment to PHYLIP
bioaln -l foo.aln # print alignment length of a CLUSTALW (default format) file
bioaln -s'100, 200' foo.aln # obtain an alignment slice
bioaln -m foo.aln # show only variable sites
bioaln -r'seq_2' foo.aln # set "seq_2" as reference (first) sequence
bioaln -g foo.aln # remove gapped sites
bioaln -p'seq_1,seq_3,seq_6' foo.aln # pick a subset of sequences
bioaln -d'seq_1,seq_3,seq_6' foo.aln # delele a subset of sequences- biotree
biotree -l foo.newick # total tree length
biotree -e 'outgroup' foo.newick # re-root on a "outgroup"
biotree -z foo.newick # print pair-wise tree distances between leafs
biotree -p 'node1' -p 'node2' foo.newick # remove "node1" and node2"
biotree -s 'node1' -s 'node2' -s 'node3' -s'node4' foo.newick # subtree consisting ONLY of these nodes- biopop
biopop --stats 'pi,theta' foo.aln # print pi and theta of population (default: CLUSTALW alignment)
biopop -m foo.aln # print mismatch distribution
biopop -d foo.aln # print distance matrix (default: Jukes-Cantor model; should be back-tracked into "bioaln")
biopop -s foo.aln # obtain number of segregating sites (i.e., SNPs)
biopop -a foo.aln # get heterozygosity for each SNP sitePower usage (with pipes)
bioseq -p'order:5' foo.fasta | bioseq -s'100,200' | bioseq -r | bioseq -t1 # pick, subseq, revcom, and translate
bioaln -o'fasta' foo.aln | bioseq -g # remove gaps within individual sequences
bioaln -o'fasta' foo.aln | bioseq -t1 | bioaln -i'fasta' # turn a nucleotide alignment into a peptide alignment
biotree -s'otu1' -s'otu2' -s'otu3' foo.newick | biotree -l # subset a large tree and get total tree lengthCreative usage (with BASH)
echo -ne ">lookup\nATG\n" | bioseq -t1 # Lookup a codon product
len=$(bioaln -l foo.aln); len_degap=$(bioaln -g foo.aln | bioaln -l); echo "$len-$len_degap" | bc -l # count alignment gaps
for i in {1..100}; do bioaln --boot foo.aln >> foo.boot; done # bootstrap an alignment (with replacement)Even-more powerful usage (with other applications)
# Permutation Trait Probability test for tree-ness:
for i in {1..100); do
bioaln -P -i'fasta' -o'fasta' nt.fas > nt.permuted.fas;
FastTree -nt nt.permuted.fas 2> /dev/null | biotree -l >> tree-length.txt # tree length of permuted alignment
done;Full documentation
POD document for bioseq
- NAME
bioseq - Fasta sequence editing module based on BioPerl.
- SYNOPSIS
bioseq [options] [sequence file]
- DESCRIPTION
bioseq will read a sequence file and act upon it by doing the following - reformat input (default is fasta) to Genbank or EMBL formats, delete specified sequences, generate overlapping subsequence with a specified window size, generate the reverese complementary sequence, for nucleic acid sequences only, take input list of sequences apart into individual sequence files, extract a specified subset of the sequence, linearize the sequence, remove gaps, find the longest open reading frame (ORF), remove stop codons, give percentage composition of specified amino acids or nuclic acid bases, split the sequences as specified by the user, translate a specific frame of input sequence, or extract a specific gene ID from multiple file sequences. By default, bioseq will assume that both the input and the output are in FASTA format.
- OPTIONS
- --help, -h
-
Print a brief help message and exit.
Usage: bioseq -h <keyword>
- --man, -m
-
Prints the manual page and exit.
Usage: bioseq -m <keyword>
- --input, -i 'format'
-
Input file format. By default, this is 'fasta'. For Genbank format, use 'genbank'. For EMBL format, use 'embl'.
Usage: bioseq -i 'genbank' input_file
- --output, -o 'format'
-
Output file format. By default, this is 'fasta'.For Genbank format, use 'genbank'. For EMBL format, use 'embl'.
Usage: bioseq -o 'EMBL' input_file
- --noseq, -n
-
Print number of sequences specified by n.
Usage: bioseq -n input_file
- --sub, -s 'min,max'
-
Select substring (of 1st sequence),
Usage: bioseq -s '<beginning index>, <ending index>' input_file Example: bioseq -s'20,80' input_file (or --sub'20,80' or -s='20,80' or --sub='20,80') - --lengths, -l
-
Print all sequence lengths.
Usage: bioseq -l
- --leadgaps, -y
-
Count and return the number of leading gaps in each sequence.
Usage: bioseq -y
- --pick, -p 'tag:value'
-
Select a single sequence or a comma-separated list of sequences, e.g, --pick 'id:foo' by id --pick 'order:2' by order --pick 're:REGEX' using a regular expression (only one regex is expected)
Specifically for a list of sequences, --pick 'id:foo,bar' list by id --pick 'order:2,3' list by order --pick 'order:2-10' list by range
Usage: bioseq -p 'id:<number>' input_file
- --del, -d 'tag:value'
- Delete a sequence or a comma-separated list of sequences, eg, --del 'id:foo' by id --del 'order:2' by order --del 'length:n' by min length, where 'n' is length --del 'ambig:x' by min % ambiguous base/aa, where 'x' is the % --del 'id:foo,bar' list by id --del 're:REGEX' using a regular expression (only one regex is expected) Usage: bioseq --del 'id:<number>' input_file
- --revcom, -r
-
Reverse complement
Usage: bioseq -r input_file
- --slidingwindow, -k 'window_size'
-
Generate overlapping subsequence with a specified window size (default 1)
Usage: bioseq -k '<index of subsequence>' input_file
- --translate, -t [1|3|6]
-
Translate in 1, 3, or 6 frames. eg, -t1, -t3, or -t6.
Usage: bioseq -t3 input_file
- --shred, -c
-
Shred into individual sequences
Usage: bioseq -c input_file
- --linearize, -L
-
Linearize FASTA.
Usage: bioseq -L fasta_file
- --extract, -e
-
Extract in-frame sequences.
Usage: bioseq -e input_file
- --nogaps, -g
-
Remove gaps
Usage: bioseq -g input_file
- --longest-orf, -C
-
Output the frame that gives the longest ORF.
Usage: bioseq -C input_file
- --removestop, -x
-
Remove stop codons (for e.g., PAML input)
Usage: bioseq -x input_file
- --anonymize, -a 'n'
-
Replace sequence IDs with serial IDs 'n' characters long, including a leading 'S' (e.g., -a'5' gives S0001). Produces a sed script file with a '.sed' suffix that may be used with sed's '-f' argument. If the filename is '-', the sed file is named STDOUT.sed instead. The sed filename is specified on STDERR.
Usage: bioseq -a <number> input_file
- --prefix 'PREFIX'
-
Used in conjunction with --anonymize. This lets you specify a custom prefix for the anonymized sequence IDs given when using the --anonymize option. If this is given, then the whole prefix will count toward the total ID length. For example: suppose the prefix chosen is SEQ, and that for --anonymize you supplied 5. Then the maximum id length is 5, so there is room for only two more digits. e.g., SEQ01 atg... SEQ02 atg... SEQ03 atc...
If there are enough sequences that the length of the prefix plus the length of the digit portion exceeds the length given to --anonymize, a warning will be given: aln-manipulations.pl -a 4 --prefix=SEQ # output SEQ1 atg... ... SEQ10 atc...
# more output WARNING: Anonymized ID length exceeded requested length: try a different length or prefix.
- --composition, -w
-
Base or AA composition.
Usage: bioseq -w input_file
- --split, -S 'split_at'
-
Split the input sequence file into several smaller files with 'split_at' sequences per file. For instance, to split a file into several smaller files with 100000 sequences each, you would run:
bioseq -S 100000 seq.fasta
The output files will be named with the label "split_N" according to the input file (or with the STDIN prefix if the file is read via standard in), where N denotes the "part" or "split" number.
- --retrieveseq, -z 'sequence retriever using GenBank accession'
- Usage: bioseq -z [Accession] Retrieves the sequence from GenBank using the provided GenBank accession. Prints out text in a fasta file. EXAMPLE: bioseq -z X83553 OUTPUT: >X83553 B.garinii (PHei strain) opsC gene. ATGAAAAAGAATACATTAAGTGCGATATTAATGACTTTATTTTTATTTATATCTTGTAAT AATTCAGGTGGGGATACTGCATCTACTAATCCTGATGAGTCTGCGAAAGGACCTAATCTT ATAGAAATAAGCAAAAAAATTACAGATTCTAATGCATTTGTACTGGCTGTGAAAGAAGTT GAGGCTTTGATCTCATCTATAGATGAACTTGCTAATAAAGCTATTGGTAAAAAAATAAAT CAAAATGGTTTAGATGCTGATGCTAATCACAACGGATCATTGTTAGCAGGAGCCCATGCA ATATCAACTCTAATAAAACAAAAAACAGATGGATTGAAAGATCTAGAAGGGTTAAGTAAA GAAATTGCAAAGGTGAAGGAATGTTCCGATAAATTTACTAAAAAGCTAACAGATAGTCAT GCACAGCTTGGAGCAGTTGGTGGTGCTATTAATGATGATCGTGCAAAAGAAGCTATTTTA AAAACACATGGGACTAACGATAAGGGTGCTAAAGAACTTAAAGAGTTATCTGAATCAGTA GAAAGCTTGGCAAAAGCAGCTCAAGCAGCATTAGCTAATTCAGTTAAAGAGCTTACAAGT CCTGTTGTGGCAGAAAGTCCAAAAAAACCTTAA
- --dotplot, -D 'draw_dotplot'
-
Extract two sequences from input file and generate a dotplot.
Usage: bioseq -D 'id1,id2,window_size,slider' fasta_file
Id1 and Id2 are extracted with their corresponding sequences. Be sure to use the entire sequence identifer, as this is a whole string match. Window_size corresponds to the number of character you would like to compare (Default window is 10). Slider is the number of windows to compare (Default slider is 10). The sequence corresponding to ID1 will appear on the X axis (row) and ID2 on the Y axis (column). This method will work on both DNA and Amino Acids.
Example: Sample Input: >id1 ATACGA >id2 ATACGA
Command: bioseq -D 'id1,id2,3' Output:
A T A C G A A * T * A * C G A A C A A T G
--rename, -R 'rename_id'
Usage: bioseq -R [file_with_new_names] file_to_be_changed.fasta
Ex: bioseq -D list.txt test-bioseq.nuc
Input:
list.txt:
VS116:7:310:IGS:11 VS116 B31:1:100:IGS:11 B31
*Left column is the pattern to be replaced by the right column
file_to_be_changed.fasta:
>VS116:7:310:IGS:11 AATTTCAAAAATATAATATAAAAACAGCTAATCCAATAGAAAAATTTGAAATTTTTCTAT TGGATAAATTCTATACAAGAAGGTAAATA >B31:1:100:IGS:11 AATTTTTAAAATATAATATAAAAACAGCTAATCCAATAGAAAAATTTTAAAACTTTTCTA TTGGATAGATTTTATACAAAGAAGGTAATA
Output: >VS116 AATTTCAAAAATATAATATAAAAACAGCTAATCCAATAGAAAAATTTGAAATTTTTCTAT TGGATAAATTCTATACAAGAAGGTAAATA >B31 AATTTTTAAAATATAATATAAAAACAGCTAATCCAATAGAAAAATTTTAAAACTTTTCTA TTGGATAGATTTTATACAAAGAAGGTAATA
- EXAMPLES
Section under construction...
- REQUIRES
Perl 5.010, BioPerl
- SEE ALSO
perl(1)
- AUTHORS
- Weigang Qiu at genectr.hunter.cuny.edu
- Yözen Hernández yzhernand at gmail dot com
- Levy Vargas levy dot vargas at gmail dot com
POD document for bioaln
POD document for biopop
POD document for biopop
Release 1.0 Notes
- Installation
- Dependency
Contributors
- Yozen Hernandez
- Levy Vargas
- Pedro Pagan
- Che Martin
- James Haven
- Girish Ramrattan
- Raymond Liang
- Saymon Akther
- Daniel Packer
- Weigang Qiu