BIOL200 2013: Difference between revisions
imported>Cmartin |
imported>Cmartin m (moved User talk:Cmartin to BIOL200 2013: rename) |
||
| (24 intermediate revisions by the same user not shown) | |||
| Line 28: | Line 28: | ||
CELL BIO II Experiment #4: | CELL BIO II Experiment #4: | ||
*'''Introduction'''<span style="font-weight:bold;color:OrangeRed;"> 1 point</span> ''':''' | *'''Introduction'''<span style="font-weight:bold;color:OrangeRed;"> 1 point</span> ''':''' | ||
<pre>Statement of objectives or aims of the experiment in the student’s own words. | |||
(not to be copied from the Lab Manual) | (not to be copied from the Lab Manual)</pre> | ||
*'''MATERIALS AND METHODS'''<span style="font-weight:bold;color:OrangeRed;"> 0 points</span> ''':''' | *'''MATERIALS AND METHODS'''<span style="font-weight:bold;color:OrangeRed;"> 0 points</span> ''':''' | ||
<pre>This should be a brief synopsis and must include any changes or deviations from the procedures | |||
outlined in the Lab Manual. Specify which organisms were used to create the phylogram.</pre> | |||
*'''RESULTS'''<span style="font-weight:bold;color:OrangeRed;"> 4 points</span> ''':''' | *'''RESULTS'''<span style="font-weight:bold;color:OrangeRed;"> 4 points</span> ''':''' | ||
<pre>A print out of the phylogram will suffice.</pre> | |||
*'''DISCUSSION'''<span style="font-weight:bold;color:OrangeRed;"> 4 points</span> ''':''' | *'''DISCUSSION'''<span style="font-weight:bold;color:OrangeRed;"> 4 points</span> ''':''' | ||
<pre>Responses to discussion questions.</pre> | |||
*'''SUMMARY |CONCLUSION'''<span style="font-weight:bold;color:OrangeRed;"> 1 point</span> ''':''' | *'''SUMMARY |CONCLUSION'''<span style="font-weight:bold;color:OrangeRed;"> 1 point</span> ''':''' | ||
<pre>Two sentence summary of your findings.</pre> | |||
*'''REFERENCES'''<span style="font-weight:bold;color:OrangeRed;"> 1 point</span> ''':''' | *'''REFERENCES'''<span style="font-weight:bold;color:OrangeRed;"> 1 point</span> ''':''' | ||
<pre>Credit is given for pertinent references obtained from sources other than the Lab Manual. | |||
This point is in addition to the 10 for the lab report.. | This point is in addition to the 10 for the lab report..</pre> | ||
===INTRODUCTION=== | ===INTRODUCTION=== | ||
| Line 51: | Line 50: | ||
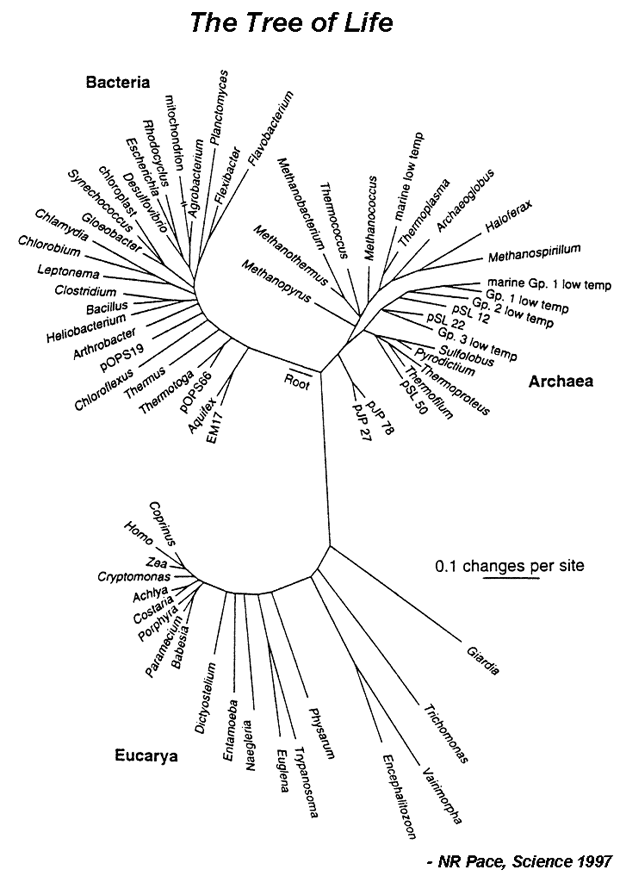
| Evolution can be defined as descent with modification. In other words, changes in the nucleotide sequence of an organsim’s genomic DNA is inherited by the next generation. According to this, all organisms are related through descent from an ancestor that lived in the distant past. Since that time, about 4 billion years ago, life has undergone an extensive process of change as new kinds of organisms arose from other kinds existing in the past.<br /> The evolutionary history of a group is called a phylogeny, and can be represented by a phylogram (Figure 1). A major goal of evolutionary analysis is to understand this history. We do not have direct knowledge of the path of evolution, as by definition, extinct organisms no longer exist. Therefore, phylogeny must be inferred indirectly. Originally, evolutionary analysis was based upon the organisms’ morphology and metabolism. This is the basis for the Linnaean classification scheme (the “Five Kingdoms” scheme). However, this method can lead to mistaken relationships. Different species living in the same environment may have similar morphologies in order to deal with specific environmental factors. Thus these similarities have nothing to do with how related the organisms are, but are a direct result of shared surroundings. However, with the advent of genomics, organisms can be grouped based upon their sequence relatedness. Since evolution is a process of inherited nucleotide change, analyzing DNA sequence differences allows for the reconstruction of a better phylogenetic history.<br/> | | Evolution can be defined as descent with modification. In other words, changes in the nucleotide sequence of an organsim’s genomic DNA is inherited by the next generation. According to this, all organisms are related through descent from an ancestor that lived in the distant past. Since that time, about 4 billion years ago, life has undergone an extensive process of change as new kinds of organisms arose from other kinds existing in the past.<br /> The evolutionary history of a group is called a phylogeny, and can be represented by a phylogram (Figure 1). A major goal of evolutionary analysis is to understand this history. We do not have direct knowledge of the path of evolution, as by definition, extinct organisms no longer exist. Therefore, phylogeny must be inferred indirectly. Originally, evolutionary analysis was based upon the organisms’ morphology and metabolism. This is the basis for the Linnaean classification scheme (the “Five Kingdoms” scheme). However, this method can lead to mistaken relationships. Different species living in the same environment may have similar morphologies in order to deal with specific environmental factors. Thus these similarities have nothing to do with how related the organisms are, but are a direct result of shared surroundings. However, with the advent of genomics, organisms can be grouped based upon their sequence relatedness. Since evolution is a process of inherited nucleotide change, analyzing DNA sequence differences allows for the reconstruction of a better phylogenetic history.<br/> | ||
|- | |- | ||
|[[File:TreeLife. | |[[File:TreeLife.PNG|center|alt=The Tree of Life.|Tree of life based on 16S ribosomal RNA (image credit: NR Pace, Science 1997)]] | ||
|-style="background-color:powderblue;" | |||
|Of course, when comparing DNA sequences, the question of which genes to use arises. The most widely used genes are those coding for the 16S rRNA gene in prokaryotes and the 18S rRNA gene in eukaryotes. These genes code for small subunit ribosomal RNA and are used for evolutionary analysis because they 1) are found in all organisms, 2) are functionally conserved, 3) vary only slightly between organisms (their nucleotide sequence changed slowly throughout evolution), and 4) have adequate length. In this lab, you will be performing evolutionary analysis by constructing a phylogram of 15 microbes spanning bacteria, archaea and eukarya. You will find and download rRNA sequences, align them and use that alignment to create a phylogram. | |Of course, when comparing DNA sequences, the question of which genes to use arises. The most widely used genes are those coding for the 16S rRNA gene in prokaryotes and the 18S rRNA gene in eukaryotes. These genes code for small subunit ribosomal RNA and are used for evolutionary analysis because they 1) are found in all organisms, 2) are functionally conserved, 3) vary only slightly between organisms (their nucleotide sequence changed slowly throughout evolution), and 4) have adequate length. In this lab, you will be performing evolutionary analysis by constructing a phylogram of 15 microbes spanning bacteria, archaea and eukarya. You will find and download rRNA sequences, align them and use that alignment to create a phylogram. | ||
|} | |} | ||
=== | ===MATERIALS=== | ||
*''' | *'''Required hardware:''' Computer | ||
{| class="collapsible collapsed wikitable" | {| class="collapsible collapsed wikitable" | ||
|- style="background-color:lightsteelblue;" | |- style="background-color:lightsteelblue;" | ||
! | ! Procedure | ||
|- style="background-color:powderblue;" | |- style="background-color:powderblue;" | ||
| | | | ||
#Examine Table I, select representative species from Bergey’s Manual. Select 2 prokaryotic species from each group, giving 14 prokaryotic species total. Also select the Eukaryotic representative, Saccharomyces cerevisiae. | |||
#Access the NCBI website: http://www.ncbi.nlm.nih.gov/ | |||
#Under the “Search” category, select “Nucleotide” | |||
#Under the “for” category, type the accession number for your first organism, and hit the “Go” button. This takes you to the access for the 16S rRNA for your organism. | |||
#Download the 16S rRNA sequence for your first organisms by choosing “FASTA” under the “Display” category. | |||
#Copy and paste the entire output into a Microsoft Word file. | |||
#Edit the sequence id to match the format of “Genus_Species_Genbank#” (eg. > Escherichia_coli_174375). | |||
#Repeat process for all of your organisms, pasting the sequences into the same Microsoft Word file. (note: be sure to place a blank line between each sequence entry) | |||
# Copy the | #Access the EMBL CLUSTALW alignment website: http://www.ebi.ac.uk/Tools/clustalw/, and copy and paste your entire Microsoft Word file into the area which asks you to “Enter or paste a set of sequences in any supported format”. Click “Run”. This program will make an alignment of all of your sequences. | ||
# | #Click “Show as Phylogram Tree” to create a tree showing the relatedness of your organisms based on their 16S rRNA sequences. | ||
# | #To print your phylogram tree.. | ||
#*a. hit the “Print Screen” button on your keyboard | |||
#*b. open the Paint program from your “accessories” menu on your computer | |||
# | #* c. hit paste to paste your screen | ||
# | #* d. “select” your phylogram tree | ||
# | #* e. copy and paste it into a new paint file | ||
# | #* f. print your tree and email it to yourself | ||
# | |||
# | |||
# | |||
# | |||
|} | |} | ||
=== | ===Table 1=== | ||
''' | {| class="wikitable" | ||
|- | |||
| colspan="2" | | |||
'''Volume 1A (Gram-negative bacteria)''' | |||
|- | |||
| | |||
''Escherichia coli'' | |||
| | |||
ACCESSION #174375 | |||
|- | |||
| | |||
''Helicobacter pylori'' | |||
| | |||
| | |||
ACCESSION #402670 | |||
|- | |||
| | |||
''Salmonella typhi'' | |||
| | |||
ACCESSION #2826789 | |||
|- | |||
| | |||
''Serratia marcescens'' | |||
| | |||
ACCESSION #4582213 | |||
|- | |||
| | |||
''Treponema pallidum'' | |||
| | |||
ACCESSION #176249 | |||
|- | |||
| colspan="2" | | |||
Additional species: ''Agrobacterium tumefaciens, Boredetella pertussis, Thermus aquaticus, Yersinia pestis, Borrelia burgdorferi. '''''(Note: To search for unlisted 16S sequences, type key words such as “yersinia<nowiki> AND 16S [gene]” in the NCBI </nowiki>GenBank search box.)''' | |||
''' | |- | ||
| colspan="2" | | |||
'''Volume 1B (Rikettsias and endosymbionts)''' | |||
|- | |||
| | |||
''Baronella bacilliformis'' | |||
| | |||
ACCESSION #173825 | |||
|- | |||
| | |||
''Chlamydia trachomatis'' | |||
| | |||
ACCESSION #2576240 | |||
|- | |||
| | |||
''Rickettsia rickettsii'' | |||
| | |||
ACCESSION #538436 | |||
|- | |||
| colspan="2" | | |||
Additional species: ''Coxiella burnetii, Thermoplasma acidophilum'' | |||
|- | |||
| colspan="2" | | |||
'''Volume 2A (Gram-positive bacteria)''' | |||
|- | |||
| | |||
''Bacillus subtilis'' | |||
| | |||
# | ACCESSION #8980302 | ||
|- | |||
| | |||
''Dinococcus radiodurans'' | |||
| | |||
ACCESSION #145033 | |||
|- | |||
| | |||
''Staphylococcus aureus'' | |||
|- | |||
| | |||
''' | |||
| | |||
ACCESSION #576603 | |||
= | |- | ||
| colspan="2" | | |||
Additional species: ''Bacillus anthracis, Clostridium botulinum, Lactobacillus acidophilus, Streptococcus pyogenes'' | |||
= | |- | ||
| colspan="2" | | |||
'''Volume 2B (Mycobacteria and nocardia)''' | |||
|- | |||
| | |||
''Mycobacterium haemophilum'' | |||
| | |||
| | |||
ACCESSION #406086 | |||
|- | |||
| | |||
''Mycobacterium tuberculosis'' | |||
|- | |||
| | |||
| | |||
ACCESSION #3929878 | |||
|- | |||
| colspan="2" | | |||
Additional species: ''Mycobacterium bovis, Nocardia orientalis'' | |||
|- | |||
| colspan="2" | | |||
'''Volume 3A (Phototrophs, chemolithotrophs, sheathed bacteria, gliding bacteria)''' | |||
|- | |||
| | |||
''Anabaena sp.'' | |||
| | |||
ACCESSION #39010 | |||
|- | |||
| | |||
''Cytophaga latercula'' | |||
| | |||
ACCESSION #37222646 | |||
|- | |||
| | |||
''Nitrobacter wiogradskyi'' | |||
| | |||
ACCESSION #402722 | |||
| | |||
= | |- | ||
'' | | colspan="2" | | ||
Additional species: ''Heliothrix oregonensis, Myxococcus fulvus, Thiobacillus ferrooxidans'' | |||
|- | |||
| colspan="2" | | |||
'''Volume 3B (Archeobaceria)''' | |||
|- | |||
| | |||
''''Methanococcus jannaschii'' | |||
|- | |||
| | |||
| | |||
ACCESSION #175446 | |||
|- | |||
| | |||
''Thermotoga subterranean'' | |||
| | |||
ACCESSION #915213 | |||
|- | |||
| colspan="2" | | |||
Additional species: ''Desulfurococcus mucosus, Halobacterium salinarium, Pyrococcus woesei'' | |||
''' | |- | ||
| colspan="2" | | |||
'''Volume 4 (Actinomycetes)''' | |||
|- | |||
| | |||
''Actinomyces bowdenii'' | |||
| | |||
ACCESSION #6456800 | |||
|- | |- | ||
| | | | ||
''' | ''Actinomyces neuii'' | ||
| | |||
ACCESSION #433527 | |||
|- | |||
| | |||
''Actinomyces turicensis'' | |||
| | |||
ACCESSION #642970 | |||
= | |- | ||
| colspan="2" | | |||
Eukaryotic representative (used as outgroup for rooting the phylogenetic tree) | |||
|- | |||
| | |||
''Saccharomyces cerevisiae'' | |||
| | |||
ACCESSION #172403 | |||
|} | |||
=== | ===ANALYSIS=== | ||
{| class="collapsible collapsed wikitable" | |||
|- style="background-color:lightsteelblue;" | |||
! Analyzing your phylogram | |||
|- style="background-color:powderblue;" | |||
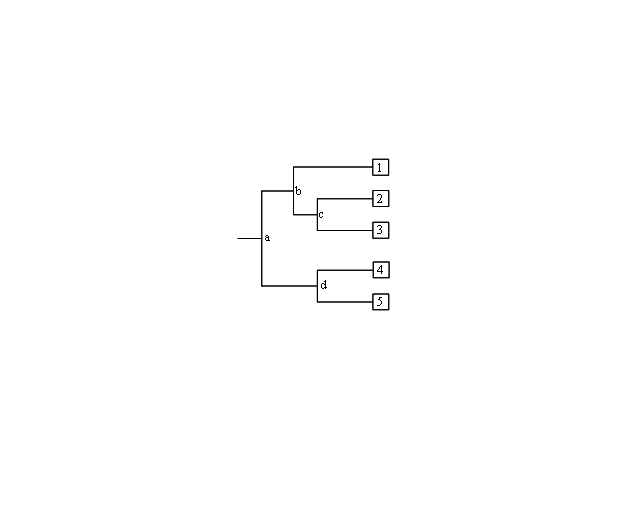
| A phyolgram is composed of nodes and branches (Figure 2). The internal nodes represent extinct ancestors, and the tips of the branches, also called nodes, are individual strains of microorganisms that exist now, and from which the sequence data were obtained. The internal nodes are points in evolution where an extinct ancestor diverged into two new entities, each of which began to accumulate differences during its subsequent independent evolution.<br/> | |||
The branches define the order of descent and the ancestry of the nodes. The branch length represents the number of changes that have occurred along that branch. Thus, the more recently two organisms share a common ancestor, the more closely related they are. Trees can be either “unrooted” or “rooted”. Unrooted trees show the relationships among the microorganisms under study, but not the evolutionary path leading from an ancestor to a strain.<br/> | |||
|- | |||
| | |||
[[ File:Phylo.PNG|center|Phylogram with internal nodes (a, b, c, d) and tips (1, 2, 3, 4, 5). Nodes at the tips are species that exist today, and internal nodes are extinct ancestors.]] | |||
|-style="background-color:powderblue;" | |||
|A rooted tree shows the unique path from an ancestor (internal node) to each strain. Trees are rooted by inclusion of an outgroup in the analysis. An outgroup is an organism that is less closely related to the other organisms under study than the organisms are to each other. | |||
|} | |||
===DISCUSSION=== | |||
{| class="collapsible collapsed wikitable" | |||
|- style="background-color:lightsteelblue;" | |||
! Discussion Questions | |||
|-style="background-color:powderblue;" | |||
| | |||
#Answer the following questions based on a Tree of Life shown in Figure 1. | |||
#*a. What do internal and terminal nodes represent? | |||
#*b. What do branch lengths represent? What’s the unit and meaning of the scale bar? | |||
#*c. Identify the positions of Humans (Homo), corn (Zea), E.coli, and Bacillus on the tree. Use the scale bar to estimate which pair is evolutionarily more distant: human/corn or E.coli/Bacillus? | |||
#In Figure 2, which two species are more closely related: 1 and 2, 2 and 3, or 1 and 4? Which are more distantly related? How did you determine this? | |||
#In Figure 2, is 1 more, less, or equally related to 4 and 5? Explain your rationale. | |||
#List and describe the key steps of constructing a phylogenetic tree. | |||
#Why do we use 18S rRNA information for yeast and 16S for prokaryotes? Could we use other molecules as phylogenetic markers? What constitutes a “good” phylogenetic marker for building a tree of life? | |||
#'''Bonus Question''' | |||
#*Define 16S “phylo-species” and “metagenomics”. Describe how PCR amplification and sequencing of 16S rRNA molecules from environmental microbial samples (e.g., sea water, soil, human gut, hot springs) can be used to define species composition of an environment. | |||
|} | |||
===References=== | |||
{| class="collapsible collapsed wikitable" | |||
|- style="background-color:lightsteelblue;" | |||
! Reference & Resource | |||
|-style="background-color:powderblue;" | |||
| | |||
#Jungck, J. R.; Fass, M.F.; Stanley, E. D. (ed.). 2003 (2006 Revision). Microbes Count! Problem Posing, Problem Solving, and Peer Persuasion in Microbiology. BioQUEST Curriculum Consortium. (Chapter 6, pg 191) | |||
#Holt. J. G. Editor-in-Chief (1984). Bergey’s Manual of Systematic Bacteriology, Volume 1-4. Williams & Wilkins: Baltimore. http://www.cme.msu.edu/bergeys/pubinfo.html | |||
|} | |||
© Weigang Qiu, Hunter College, Last Update Jan 2013 | © Weigang Qiu, Hunter College, Last Update Jan 2013 | ||
Latest revision as of 20:38, 4 March 2013
EXPERIMENT # 4
BIOL 200 Cell Biology II LAB, Spring 2013
Hunter College of the City University of New York
Course information
Instructors: TBD
Class Hours: Room TBD HN; TBD
Office Hours: Room 830 HN; Thursdays 2-4pm or by appointment
Contact information:
- Dr. Weigang Qiu: weigang@genectr.hunter.cuny.edu, 1-212-772-5296
Experiment #4
The Tree of Life and Molecular Identification of Microorganisms
Objective
To classify microorganisms and determine their relatedness using molecular sequences.
LAB REPORT GRADING GUIDE
CELL BIO II Experiment #4:
- Introduction 1 point :
Statement of objectives or aims of the experiment in the student’s own words. (not to be copied from the Lab Manual)
- MATERIALS AND METHODS 0 points :
This should be a brief synopsis and must include any changes or deviations from the procedures outlined in the Lab Manual. Specify which organisms were used to create the phylogram.
- RESULTS 4 points :
A print out of the phylogram will suffice.
- DISCUSSION 4 points :
Responses to discussion questions.
- SUMMARY |CONCLUSION 1 point :
Two sentence summary of your findings.
- REFERENCES 1 point :
Credit is given for pertinent references obtained from sources other than the Lab Manual. This point is in addition to the 10 for the lab report..
INTRODUCTION
| Introduction |
|---|
| Evolution can be defined as descent with modification. In other words, changes in the nucleotide sequence of an organsim’s genomic DNA is inherited by the next generation. According to this, all organisms are related through descent from an ancestor that lived in the distant past. Since that time, about 4 billion years ago, life has undergone an extensive process of change as new kinds of organisms arose from other kinds existing in the past. The evolutionary history of a group is called a phylogeny, and can be represented by a phylogram (Figure 1). A major goal of evolutionary analysis is to understand this history. We do not have direct knowledge of the path of evolution, as by definition, extinct organisms no longer exist. Therefore, phylogeny must be inferred indirectly. Originally, evolutionary analysis was based upon the organisms’ morphology and metabolism. This is the basis for the Linnaean classification scheme (the “Five Kingdoms” scheme). However, this method can lead to mistaken relationships. Different species living in the same environment may have similar morphologies in order to deal with specific environmental factors. Thus these similarities have nothing to do with how related the organisms are, but are a direct result of shared surroundings. However, with the advent of genomics, organisms can be grouped based upon their sequence relatedness. Since evolution is a process of inherited nucleotide change, analyzing DNA sequence differences allows for the reconstruction of a better phylogenetic history. |
| Of course, when comparing DNA sequences, the question of which genes to use arises. The most widely used genes are those coding for the 16S rRNA gene in prokaryotes and the 18S rRNA gene in eukaryotes. These genes code for small subunit ribosomal RNA and are used for evolutionary analysis because they 1) are found in all organisms, 2) are functionally conserved, 3) vary only slightly between organisms (their nucleotide sequence changed slowly throughout evolution), and 4) have adequate length. In this lab, you will be performing evolutionary analysis by constructing a phylogram of 15 microbes spanning bacteria, archaea and eukarya. You will find and download rRNA sequences, align them and use that alignment to create a phylogram. |
MATERIALS
- Required hardware: Computer
| Procedure |
|---|
|
Table 1
|
Volume 1A (Gram-negative bacteria) | |
|
Escherichia coli |
ACCESSION #174375 |
|
Helicobacter pylori |
ACCESSION #402670 |
|
Salmonella typhi |
ACCESSION #2826789 |
|
Serratia marcescens |
ACCESSION #4582213 |
|
Treponema pallidum |
ACCESSION #176249 |
|
Additional species: Agrobacterium tumefaciens, Boredetella pertussis, Thermus aquaticus, Yersinia pestis, Borrelia burgdorferi. (Note: To search for unlisted 16S sequences, type key words such as “yersinia AND 16S [gene]” in the NCBI GenBank search box.) | |
|
Volume 1B (Rikettsias and endosymbionts) | |
|
Baronella bacilliformis |
ACCESSION #173825 |
|
Chlamydia trachomatis |
ACCESSION #2576240 |
|
Rickettsia rickettsii |
ACCESSION #538436 |
|
Additional species: Coxiella burnetii, Thermoplasma acidophilum | |
|
Volume 2A (Gram-positive bacteria) | |
|
Bacillus subtilis |
ACCESSION #8980302 |
|
Dinococcus radiodurans |
ACCESSION #145033 |
|
Staphylococcus aureus |
ACCESSION #576603 |
|
Additional species: Bacillus anthracis, Clostridium botulinum, Lactobacillus acidophilus, Streptococcus pyogenes | |
|
Volume 2B (Mycobacteria and nocardia) | |
|
Mycobacterium haemophilum |
ACCESSION #406086 |
|
Mycobacterium tuberculosis |
ACCESSION #3929878 |
|
Additional species: Mycobacterium bovis, Nocardia orientalis | |
|
Volume 3A (Phototrophs, chemolithotrophs, sheathed bacteria, gliding bacteria) | |
|
Anabaena sp. |
ACCESSION #39010 |
|
Cytophaga latercula |
ACCESSION #37222646 |
|
Nitrobacter wiogradskyi |
ACCESSION #402722 |
|
Additional species: Heliothrix oregonensis, Myxococcus fulvus, Thiobacillus ferrooxidans | |
|
Volume 3B (Archeobaceria) | |
|
''Methanococcus jannaschii |
ACCESSION #175446 |
|
Thermotoga subterranean |
ACCESSION #915213 |
|
Additional species: Desulfurococcus mucosus, Halobacterium salinarium, Pyrococcus woesei | |
|
Volume 4 (Actinomycetes) | |
|
Actinomyces bowdenii |
ACCESSION #6456800 |
|
Actinomyces neuii |
ACCESSION #433527 |
|
Actinomyces turicensis |
ACCESSION #642970 |
|
Eukaryotic representative (used as outgroup for rooting the phylogenetic tree) | |
|
Saccharomyces cerevisiae |
ACCESSION #172403 |
ANALYSIS
| Analyzing your phylogram |
|---|
| A phyolgram is composed of nodes and branches (Figure 2). The internal nodes represent extinct ancestors, and the tips of the branches, also called nodes, are individual strains of microorganisms that exist now, and from which the sequence data were obtained. The internal nodes are points in evolution where an extinct ancestor diverged into two new entities, each of which began to accumulate differences during its subsequent independent evolution. The branches define the order of descent and the ancestry of the nodes. The branch length represents the number of changes that have occurred along that branch. Thus, the more recently two organisms share a common ancestor, the more closely related they are. Trees can be either “unrooted” or “rooted”. Unrooted trees show the relationships among the microorganisms under study, but not the evolutionary path leading from an ancestor to a strain. |
| A rooted tree shows the unique path from an ancestor (internal node) to each strain. Trees are rooted by inclusion of an outgroup in the analysis. An outgroup is an organism that is less closely related to the other organisms under study than the organisms are to each other. |
DISCUSSION
| Discussion Questions |
|---|
|
References
| Reference & Resource |
|---|
|
© Weigang Qiu, Hunter College, Last Update Jan 2013